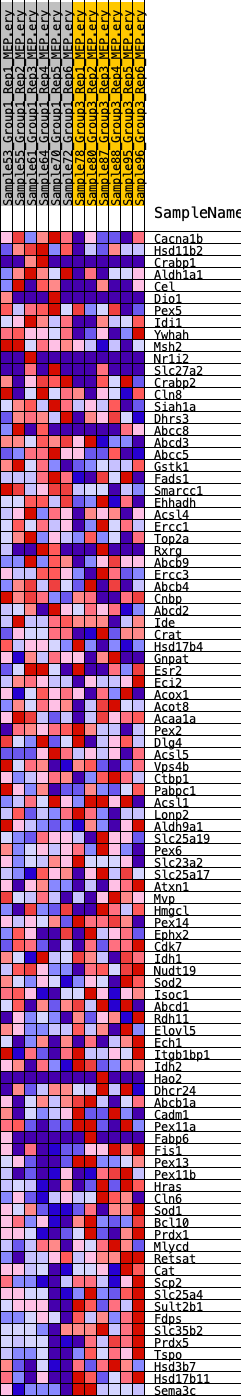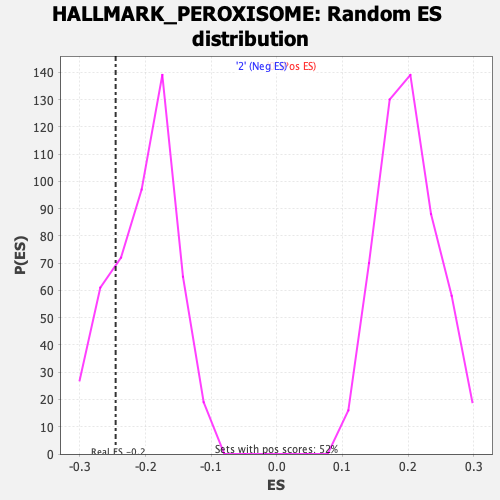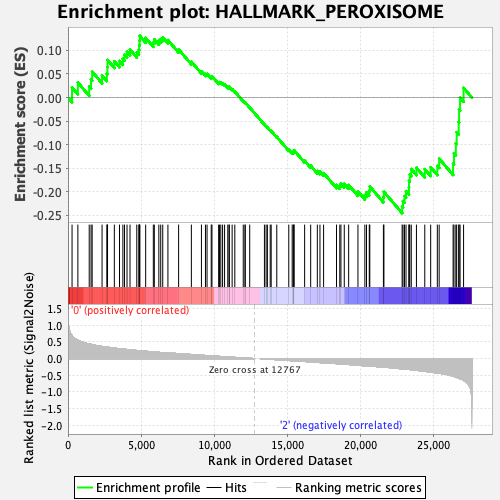
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group3.MEP.ery_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.2455122 |
| Normalized Enrichment Score (NES) | -1.2078613 |
| Nominal p-value | 0.2125 |
| FDR q-value | 0.56547046 |
| FWER p-Value | 0.933 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cacna1b | 277 | 0.687 | 0.0212 | No |
| 2 | Hsd11b2 | 675 | 0.548 | 0.0318 | No |
| 3 | Crabp1 | 1450 | 0.435 | 0.0235 | No |
| 4 | Aldh1a1 | 1576 | 0.427 | 0.0384 | No |
| 5 | Cel | 1651 | 0.421 | 0.0549 | No |
| 6 | Dio1 | 2328 | 0.364 | 0.0469 | No |
| 7 | Pex5 | 2656 | 0.341 | 0.0505 | No |
| 8 | Idi1 | 2693 | 0.338 | 0.0646 | No |
| 9 | Ywhah | 2702 | 0.337 | 0.0797 | No |
| 10 | Msh2 | 3167 | 0.314 | 0.0771 | No |
| 11 | Nr1i2 | 3521 | 0.294 | 0.0777 | No |
| 12 | Slc27a2 | 3751 | 0.282 | 0.0822 | No |
| 13 | Crabp2 | 3861 | 0.280 | 0.0910 | No |
| 14 | Cln8 | 4034 | 0.272 | 0.0971 | No |
| 15 | Siah1a | 4239 | 0.262 | 0.1017 | No |
| 16 | Dhrs3 | 4702 | 0.242 | 0.0959 | No |
| 17 | Abcc8 | 4846 | 0.235 | 0.1014 | No |
| 18 | Abcd3 | 4863 | 0.234 | 0.1115 | No |
| 19 | Abcc5 | 4894 | 0.232 | 0.1210 | No |
| 20 | Gstk1 | 4910 | 0.232 | 0.1310 | No |
| 21 | Fads1 | 5309 | 0.221 | 0.1266 | No |
| 22 | Smarcc1 | 5832 | 0.201 | 0.1167 | No |
| 23 | Ehhadh | 5900 | 0.199 | 0.1234 | No |
| 24 | Acsl4 | 6209 | 0.186 | 0.1207 | No |
| 25 | Ercc1 | 6338 | 0.182 | 0.1243 | No |
| 26 | Top2a | 6475 | 0.177 | 0.1274 | No |
| 27 | Rxrg | 6830 | 0.165 | 0.1221 | No |
| 28 | Abcb9 | 7561 | 0.148 | 0.1022 | No |
| 29 | Ercc3 | 8432 | 0.121 | 0.0761 | No |
| 30 | Abcb4 | 9121 | 0.101 | 0.0557 | No |
| 31 | Cnbp | 9395 | 0.093 | 0.0500 | No |
| 32 | Abcd2 | 9504 | 0.090 | 0.0502 | No |
| 33 | Ide | 9790 | 0.081 | 0.0435 | No |
| 34 | Crat | 9840 | 0.079 | 0.0454 | No |
| 35 | Hsd17b4 | 10297 | 0.067 | 0.0318 | No |
| 36 | Gnpat | 10353 | 0.065 | 0.0328 | No |
| 37 | Esr2 | 10445 | 0.062 | 0.0323 | No |
| 38 | Eci2 | 10566 | 0.059 | 0.0306 | No |
| 39 | Acox1 | 10703 | 0.055 | 0.0282 | No |
| 40 | Acot8 | 10923 | 0.048 | 0.0224 | No |
| 41 | Acaa1a | 11017 | 0.045 | 0.0211 | No |
| 42 | Pex2 | 11019 | 0.045 | 0.0231 | No |
| 43 | Dlg4 | 11219 | 0.040 | 0.0177 | No |
| 44 | Acsl5 | 11402 | 0.035 | 0.0127 | No |
| 45 | Vps4b | 11979 | 0.021 | -0.0073 | No |
| 46 | Ctbp1 | 12098 | 0.018 | -0.0107 | No |
| 47 | Pabpc1 | 12115 | 0.018 | -0.0105 | No |
| 48 | Acsl1 | 12421 | 0.009 | -0.0212 | No |
| 49 | Lonp2 | 13427 | -0.008 | -0.0574 | No |
| 50 | Aldh9a1 | 13540 | -0.011 | -0.0609 | No |
| 51 | Slc25a19 | 13629 | -0.013 | -0.0635 | No |
| 52 | Pex6 | 13821 | -0.019 | -0.0696 | No |
| 53 | Slc23a2 | 13888 | -0.021 | -0.0711 | No |
| 54 | Slc25a17 | 14271 | -0.032 | -0.0835 | No |
| 55 | Atxn1 | 15091 | -0.055 | -0.1108 | No |
| 56 | Mvp | 15324 | -0.063 | -0.1163 | No |
| 57 | Hmgcl | 15403 | -0.065 | -0.1162 | No |
| 58 | Pex14 | 15444 | -0.066 | -0.1147 | No |
| 59 | Ephx2 | 15446 | -0.066 | -0.1117 | No |
| 60 | Cdk7 | 16175 | -0.086 | -0.1342 | No |
| 61 | Idh1 | 16588 | -0.099 | -0.1447 | No |
| 62 | Nudt19 | 17042 | -0.113 | -0.1560 | No |
| 63 | Sod2 | 17222 | -0.119 | -0.1571 | No |
| 64 | Isoc1 | 17471 | -0.124 | -0.1605 | No |
| 65 | Abcd1 | 18354 | -0.149 | -0.1858 | No |
| 66 | Rdh11 | 18578 | -0.157 | -0.1867 | No |
| 67 | Elovl5 | 18656 | -0.159 | -0.1823 | No |
| 68 | Ech1 | 18884 | -0.167 | -0.1830 | No |
| 69 | Itgb1bp1 | 19184 | -0.176 | -0.1858 | No |
| 70 | Idh2 | 19819 | -0.200 | -0.1997 | No |
| 71 | Hao2 | 20292 | -0.216 | -0.2071 | No |
| 72 | Dhcr24 | 20404 | -0.216 | -0.2012 | No |
| 73 | Abcb1a | 20593 | -0.224 | -0.1979 | No |
| 74 | Cadm1 | 20630 | -0.225 | -0.1889 | No |
| 75 | Pex11a | 21559 | -0.259 | -0.2109 | No |
| 76 | Fabp6 | 21594 | -0.260 | -0.2003 | No |
| 77 | Fis1 | 22839 | -0.303 | -0.2317 | Yes |
| 78 | Pex13 | 22898 | -0.306 | -0.2199 | Yes |
| 79 | Pex11b | 22999 | -0.311 | -0.2093 | Yes |
| 80 | Hras | 23105 | -0.315 | -0.1988 | Yes |
| 81 | Cln6 | 23305 | -0.325 | -0.1912 | Yes |
| 82 | Sod1 | 23311 | -0.326 | -0.1766 | Yes |
| 83 | Bcl10 | 23362 | -0.327 | -0.1635 | Yes |
| 84 | Prdx1 | 23464 | -0.333 | -0.1520 | Yes |
| 85 | Mlycd | 23817 | -0.349 | -0.1489 | Yes |
| 86 | Retsat | 24387 | -0.384 | -0.1521 | Yes |
| 87 | Cat | 24790 | -0.406 | -0.1482 | Yes |
| 88 | Scp2 | 25245 | -0.428 | -0.1452 | Yes |
| 89 | Slc25a4 | 25370 | -0.435 | -0.1298 | Yes |
| 90 | Sult2b1 | 26324 | -0.528 | -0.1404 | Yes |
| 91 | Fdps | 26383 | -0.535 | -0.1181 | Yes |
| 92 | Slc35b2 | 26511 | -0.557 | -0.0974 | Yes |
| 93 | Prdx5 | 26567 | -0.565 | -0.0736 | Yes |
| 94 | Tspo | 26704 | -0.589 | -0.0517 | Yes |
| 95 | Hsd3b7 | 26735 | -0.596 | -0.0257 | Yes |
| 96 | Hsd17b11 | 26800 | -0.605 | -0.0004 | Yes |
| 97 | Sema3c | 27039 | -0.648 | 0.0205 | Yes |