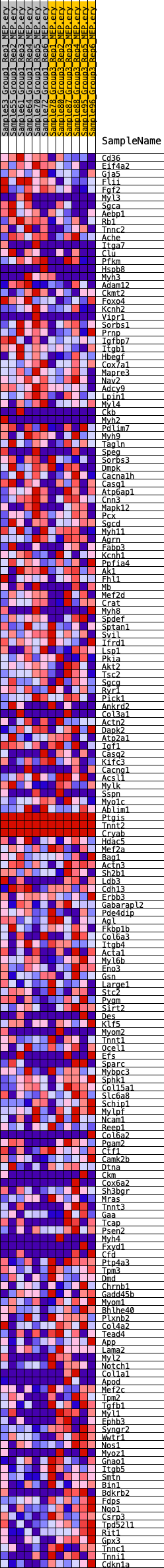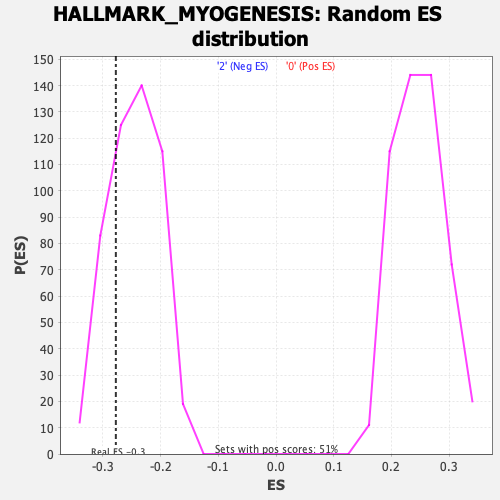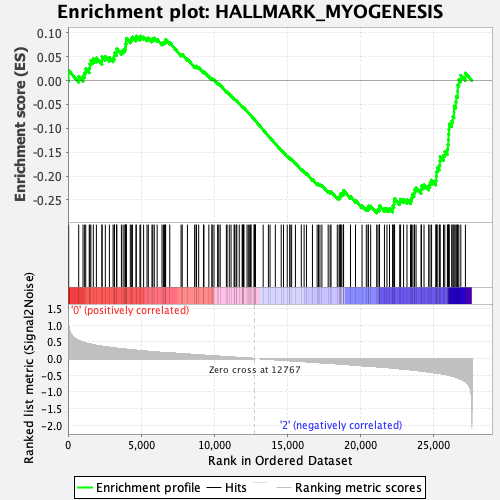
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group3.MEP.ery_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.27754584 |
| Normalized Enrichment Score (NES) | -1.1294142 |
| Nominal p-value | 0.24696356 |
| FDR q-value | 0.5439711 |
| FWER p-Value | 0.971 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cd36 | 46 | 0.974 | 0.0211 | No |
| 2 | Eif4a2 | 731 | 0.539 | 0.0088 | No |
| 3 | Gja5 | 1026 | 0.491 | 0.0095 | No |
| 4 | Flii | 1136 | 0.473 | 0.0166 | No |
| 5 | Fgf2 | 1212 | 0.464 | 0.0248 | No |
| 6 | Myl3 | 1452 | 0.435 | 0.0262 | No |
| 7 | Sgca | 1497 | 0.432 | 0.0347 | No |
| 8 | Aebp1 | 1561 | 0.429 | 0.0425 | No |
| 9 | Rb1 | 1730 | 0.411 | 0.0460 | No |
| 10 | Tnnc2 | 1940 | 0.392 | 0.0475 | No |
| 11 | Ache | 2322 | 0.364 | 0.0421 | No |
| 12 | Itga7 | 2325 | 0.364 | 0.0506 | No |
| 13 | Clu | 2552 | 0.350 | 0.0505 | No |
| 14 | Pfkm | 2829 | 0.335 | 0.0483 | No |
| 15 | Hspb8 | 3081 | 0.318 | 0.0466 | No |
| 16 | Myh3 | 3162 | 0.315 | 0.0510 | No |
| 17 | Adam12 | 3176 | 0.313 | 0.0579 | No |
| 18 | Ckmt2 | 3327 | 0.305 | 0.0595 | No |
| 19 | Foxo4 | 3333 | 0.305 | 0.0665 | No |
| 20 | Kcnh2 | 3651 | 0.288 | 0.0617 | No |
| 21 | Vipr1 | 3767 | 0.281 | 0.0641 | No |
| 22 | Sorbs1 | 3886 | 0.279 | 0.0663 | No |
| 23 | Prnp | 3934 | 0.277 | 0.0711 | No |
| 24 | Igfbp7 | 3937 | 0.277 | 0.0775 | No |
| 25 | Itgb1 | 3982 | 0.274 | 0.0823 | No |
| 26 | Hbegf | 3986 | 0.274 | 0.0886 | No |
| 27 | Cox7a1 | 4258 | 0.261 | 0.0848 | No |
| 28 | Mapre3 | 4329 | 0.258 | 0.0883 | No |
| 29 | Nav2 | 4405 | 0.255 | 0.0915 | No |
| 30 | Adcy9 | 4650 | 0.244 | 0.0883 | No |
| 31 | Lpin1 | 4665 | 0.244 | 0.0935 | No |
| 32 | Myl4 | 4917 | 0.232 | 0.0898 | No |
| 33 | Ckb | 4969 | 0.229 | 0.0933 | No |
| 34 | Myh2 | 5168 | 0.225 | 0.0914 | No |
| 35 | Pdlim7 | 5401 | 0.217 | 0.0880 | No |
| 36 | Myh9 | 5490 | 0.213 | 0.0897 | No |
| 37 | Tagln | 5740 | 0.205 | 0.0855 | No |
| 38 | Speg | 5796 | 0.203 | 0.0882 | No |
| 39 | Sorbs3 | 5898 | 0.199 | 0.0892 | No |
| 40 | Dmpk | 6094 | 0.191 | 0.0865 | No |
| 41 | Cacna1h | 6426 | 0.179 | 0.0786 | No |
| 42 | Casq1 | 6522 | 0.175 | 0.0793 | No |
| 43 | Atp6ap1 | 6592 | 0.173 | 0.0808 | No |
| 44 | Cnn3 | 6667 | 0.171 | 0.0821 | No |
| 45 | Mapk12 | 6671 | 0.171 | 0.0860 | No |
| 46 | Pcx | 6955 | 0.161 | 0.0794 | No |
| 47 | Sgcd | 7729 | 0.142 | 0.0546 | No |
| 48 | Myh11 | 7808 | 0.140 | 0.0550 | No |
| 49 | Agrn | 8162 | 0.130 | 0.0452 | No |
| 50 | Fabp3 | 8655 | 0.114 | 0.0299 | No |
| 51 | Kcnh1 | 8754 | 0.112 | 0.0289 | No |
| 52 | Ppfia4 | 8785 | 0.110 | 0.0304 | No |
| 53 | Ak1 | 8932 | 0.106 | 0.0276 | No |
| 54 | Fhl1 | 9259 | 0.097 | 0.0180 | No |
| 55 | Mb | 9293 | 0.096 | 0.0190 | No |
| 56 | Mef2d | 9627 | 0.085 | 0.0089 | No |
| 57 | Crat | 9840 | 0.079 | 0.0030 | No |
| 58 | Myh8 | 9849 | 0.079 | 0.0046 | No |
| 59 | Spdef | 9980 | 0.076 | 0.0016 | No |
| 60 | Sptan1 | 10225 | 0.069 | -0.0057 | No |
| 61 | Svil | 10306 | 0.067 | -0.0071 | No |
| 62 | Ifrd1 | 10402 | 0.063 | -0.0090 | No |
| 63 | Lsp1 | 10825 | 0.051 | -0.0232 | No |
| 64 | Pkia | 10873 | 0.050 | -0.0238 | No |
| 65 | Akt2 | 11024 | 0.045 | -0.0282 | No |
| 66 | Tsc2 | 11145 | 0.042 | -0.0316 | No |
| 67 | Sgcg | 11351 | 0.036 | -0.0382 | No |
| 68 | Ryr1 | 11396 | 0.035 | -0.0390 | No |
| 69 | Pick1 | 11493 | 0.032 | -0.0417 | No |
| 70 | Ankrd2 | 11539 | 0.031 | -0.0427 | No |
| 71 | Col3a1 | 11705 | 0.027 | -0.0480 | No |
| 72 | Actn2 | 11903 | 0.023 | -0.0547 | No |
| 73 | Dapk2 | 11940 | 0.022 | -0.0555 | No |
| 74 | Atp2a1 | 11957 | 0.022 | -0.0555 | No |
| 75 | Igf1 | 11997 | 0.021 | -0.0565 | No |
| 76 | Casq2 | 12028 | 0.020 | -0.0571 | No |
| 77 | Kifc3 | 12237 | 0.014 | -0.0643 | No |
| 78 | Cacng1 | 12324 | 0.011 | -0.0672 | No |
| 79 | Acsl1 | 12421 | 0.009 | -0.0705 | No |
| 80 | Mylk | 12463 | 0.007 | -0.0718 | No |
| 81 | Sspn | 12518 | 0.006 | -0.0737 | No |
| 82 | Myo1c | 12712 | 0.002 | -0.0807 | No |
| 83 | Ablim1 | 12753 | 0.000 | -0.0821 | No |
| 84 | Ptgis | 12774 | 0.000 | -0.0828 | No |
| 85 | Tnnt2 | 12795 | 0.000 | -0.0836 | No |
| 86 | Cryab | 12811 | 0.000 | -0.0841 | No |
| 87 | Hdac5 | 13341 | -0.005 | -0.1033 | No |
| 88 | Mef2a | 13697 | -0.016 | -0.1159 | No |
| 89 | Bag1 | 13810 | -0.018 | -0.1195 | No |
| 90 | Actn3 | 14172 | -0.029 | -0.1320 | No |
| 91 | Sh2b1 | 14574 | -0.040 | -0.1457 | No |
| 92 | Ldb3 | 14730 | -0.045 | -0.1503 | No |
| 93 | Cdh13 | 14986 | -0.052 | -0.1584 | No |
| 94 | Erbb3 | 15167 | -0.058 | -0.1636 | No |
| 95 | Gabarapl2 | 15168 | -0.058 | -0.1622 | No |
| 96 | Pde4dip | 15287 | -0.062 | -0.1651 | No |
| 97 | Agl | 15545 | -0.069 | -0.1728 | No |
| 98 | Fkbp1b | 15946 | -0.081 | -0.1855 | No |
| 99 | Col6a3 | 16140 | -0.085 | -0.1906 | No |
| 100 | Itgb4 | 16299 | -0.089 | -0.1943 | No |
| 101 | Acta1 | 16709 | -0.102 | -0.2068 | No |
| 102 | Myl6b | 17029 | -0.112 | -0.2158 | No |
| 103 | Eno3 | 17125 | -0.116 | -0.2165 | No |
| 104 | Gsn | 17221 | -0.119 | -0.2172 | No |
| 105 | Large1 | 17355 | -0.120 | -0.2193 | No |
| 106 | Stc2 | 17784 | -0.134 | -0.2317 | No |
| 107 | Pygm | 17915 | -0.138 | -0.2333 | No |
| 108 | Sirt2 | 17979 | -0.140 | -0.2323 | No |
| 109 | Des | 18407 | -0.151 | -0.2443 | No |
| 110 | Klf5 | 18529 | -0.155 | -0.2451 | No |
| 111 | Myom2 | 18590 | -0.157 | -0.2436 | No |
| 112 | Tnnt1 | 18610 | -0.157 | -0.2406 | No |
| 113 | Ocel1 | 18613 | -0.157 | -0.2370 | No |
| 114 | Efs | 18707 | -0.161 | -0.2367 | No |
| 115 | Sparc | 18822 | -0.165 | -0.2370 | No |
| 116 | Mybpc3 | 18825 | -0.165 | -0.2332 | No |
| 117 | Sphk1 | 18837 | -0.166 | -0.2297 | No |
| 118 | Col15a1 | 19310 | -0.181 | -0.2427 | No |
| 119 | Slc6a8 | 19652 | -0.194 | -0.2506 | No |
| 120 | Schip1 | 20098 | -0.209 | -0.2619 | No |
| 121 | Mylpf | 20388 | -0.216 | -0.2674 | No |
| 122 | Ncam1 | 20512 | -0.221 | -0.2667 | No |
| 123 | Reep1 | 20531 | -0.221 | -0.2622 | No |
| 124 | Col6a2 | 20689 | -0.227 | -0.2626 | No |
| 125 | Pgam2 | 21100 | -0.239 | -0.2719 | Yes |
| 126 | Ctf1 | 21190 | -0.242 | -0.2695 | Yes |
| 127 | Camk2b | 21288 | -0.247 | -0.2673 | Yes |
| 128 | Dtna | 21293 | -0.247 | -0.2616 | Yes |
| 129 | Ckm | 21630 | -0.260 | -0.2678 | Yes |
| 130 | Cox6a2 | 21802 | -0.265 | -0.2678 | Yes |
| 131 | Sh3bgr | 21976 | -0.270 | -0.2678 | Yes |
| 132 | Mras | 22194 | -0.279 | -0.2692 | Yes |
| 133 | Tnnt3 | 22200 | -0.280 | -0.2628 | Yes |
| 134 | Gaa | 22278 | -0.283 | -0.2590 | Yes |
| 135 | Tcap | 22291 | -0.283 | -0.2528 | Yes |
| 136 | Psen2 | 22323 | -0.284 | -0.2473 | Yes |
| 137 | Myh4 | 22675 | -0.300 | -0.2531 | Yes |
| 138 | Fxyd1 | 22734 | -0.300 | -0.2482 | Yes |
| 139 | Cfd | 22938 | -0.309 | -0.2484 | Yes |
| 140 | Ptp4a3 | 23171 | -0.319 | -0.2494 | Yes |
| 141 | Tpm3 | 23413 | -0.330 | -0.2504 | Yes |
| 142 | Dmd | 23483 | -0.333 | -0.2451 | Yes |
| 143 | Chrnb1 | 23525 | -0.335 | -0.2388 | Yes |
| 144 | Gadd45b | 23655 | -0.340 | -0.2355 | Yes |
| 145 | Myom1 | 23688 | -0.341 | -0.2287 | Yes |
| 146 | Bhlhe40 | 23792 | -0.347 | -0.2243 | Yes |
| 147 | Plxnb2 | 24133 | -0.368 | -0.2281 | Yes |
| 148 | Col4a2 | 24159 | -0.370 | -0.2204 | Yes |
| 149 | Tead4 | 24336 | -0.381 | -0.2179 | Yes |
| 150 | App | 24654 | -0.396 | -0.2202 | Yes |
| 151 | Lama2 | 24743 | -0.403 | -0.2139 | Yes |
| 152 | Myl2 | 24852 | -0.409 | -0.2083 | Yes |
| 153 | Notch1 | 25142 | -0.421 | -0.2090 | Yes |
| 154 | Col1a1 | 25171 | -0.424 | -0.2001 | Yes |
| 155 | Apod | 25189 | -0.424 | -0.1908 | Yes |
| 156 | Mef2c | 25237 | -0.428 | -0.1825 | Yes |
| 157 | Tpm2 | 25374 | -0.436 | -0.1773 | Yes |
| 158 | Tgfb1 | 25418 | -0.440 | -0.1685 | Yes |
| 159 | Myl1 | 25434 | -0.441 | -0.1588 | Yes |
| 160 | Ephb3 | 25669 | -0.461 | -0.1565 | Yes |
| 161 | Syngr2 | 25755 | -0.468 | -0.1487 | Yes |
| 162 | Wwtr1 | 25933 | -0.481 | -0.1438 | Yes |
| 163 | Nos1 | 25965 | -0.485 | -0.1336 | Yes |
| 164 | Myoz1 | 26002 | -0.488 | -0.1235 | Yes |
| 165 | Gnao1 | 26014 | -0.490 | -0.1125 | Yes |
| 166 | Itgb5 | 26037 | -0.492 | -0.1017 | Yes |
| 167 | Smtn | 26054 | -0.495 | -0.0907 | Yes |
| 168 | Bin1 | 26224 | -0.514 | -0.0849 | Yes |
| 169 | Bdkrb2 | 26304 | -0.525 | -0.0755 | Yes |
| 170 | Fdps | 26383 | -0.535 | -0.0658 | Yes |
| 171 | Nqo1 | 26388 | -0.536 | -0.0534 | Yes |
| 172 | Csrp3 | 26512 | -0.557 | -0.0448 | Yes |
| 173 | Tpd52l1 | 26539 | -0.561 | -0.0326 | Yes |
| 174 | Rit1 | 26634 | -0.575 | -0.0226 | Yes |
| 175 | Gpx3 | 26642 | -0.577 | -0.0094 | Yes |
| 176 | Tnnc1 | 26717 | -0.592 | 0.0018 | Yes |
| 177 | Tnni1 | 26851 | -0.614 | 0.0113 | Yes |
| 178 | Cdkn1a | 27165 | -0.686 | 0.0159 | Yes |