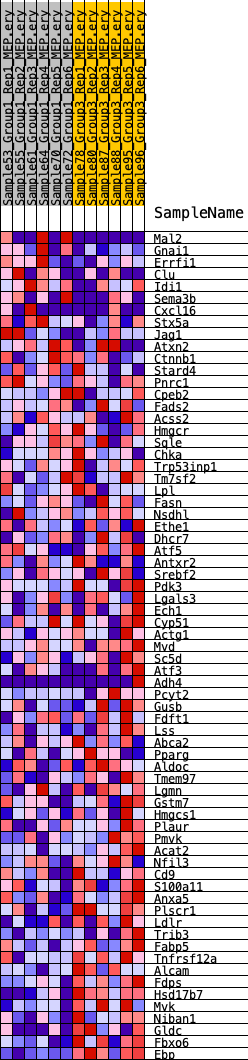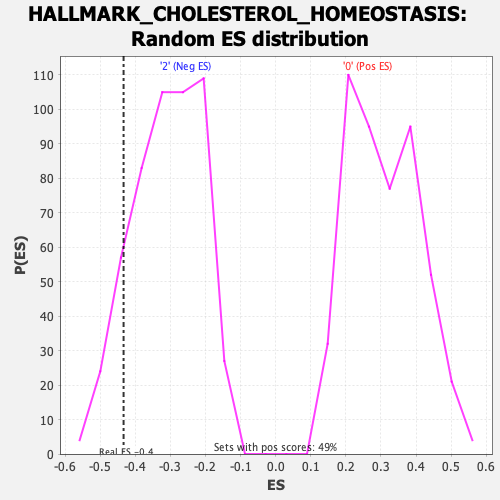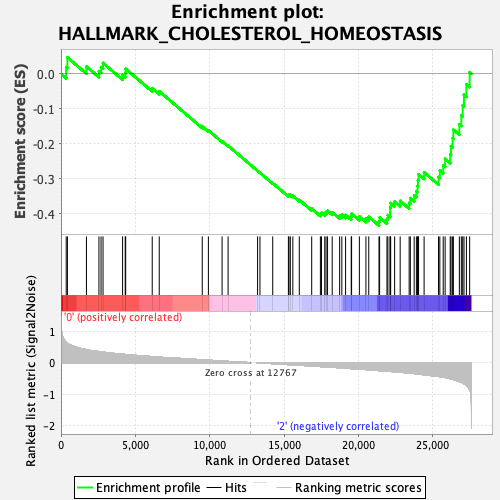
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group3.MEP.ery_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.43313769 |
| Normalized Enrichment Score (NES) | -1.4002382 |
| Nominal p-value | 0.10311284 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.706 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mal2 | 350 | 0.644 | 0.0191 | No |
| 2 | Gnai1 | 431 | 0.617 | 0.0467 | No |
| 3 | Errfi1 | 1710 | 0.413 | 0.0207 | No |
| 4 | Clu | 2552 | 0.350 | 0.0074 | No |
| 5 | Idi1 | 2693 | 0.338 | 0.0190 | No |
| 6 | Sema3b | 2826 | 0.335 | 0.0308 | No |
| 7 | Cxcl16 | 4142 | 0.267 | -0.0038 | No |
| 8 | Stx5a | 4319 | 0.259 | 0.0026 | No |
| 9 | Jag1 | 4349 | 0.257 | 0.0142 | No |
| 10 | Atxn2 | 6134 | 0.189 | -0.0412 | No |
| 11 | Ctnnb1 | 6606 | 0.173 | -0.0498 | No |
| 12 | Stard4 | 9498 | 0.090 | -0.1504 | No |
| 13 | Pnrc1 | 9911 | 0.077 | -0.1615 | No |
| 14 | Cpeb2 | 10832 | 0.051 | -0.1924 | No |
| 15 | Fads2 | 11236 | 0.040 | -0.2051 | No |
| 16 | Acss2 | 13219 | -0.002 | -0.2770 | No |
| 17 | Hmgcr | 13378 | -0.006 | -0.2824 | No |
| 18 | Sqle | 14237 | -0.031 | -0.3120 | No |
| 19 | Chka | 15291 | -0.062 | -0.3472 | No |
| 20 | Trp53inp1 | 15329 | -0.063 | -0.3455 | No |
| 21 | Tm7sf2 | 15420 | -0.066 | -0.3455 | No |
| 22 | Lpl | 15589 | -0.070 | -0.3481 | No |
| 23 | Fasn | 16023 | -0.082 | -0.3598 | No |
| 24 | Nsdhl | 16858 | -0.107 | -0.3848 | No |
| 25 | Ethe1 | 17442 | -0.123 | -0.3999 | No |
| 26 | Dhcr7 | 17516 | -0.125 | -0.3963 | No |
| 27 | Atf5 | 17736 | -0.133 | -0.3977 | No |
| 28 | Antxr2 | 17823 | -0.135 | -0.3942 | No |
| 29 | Srebf2 | 17924 | -0.138 | -0.3910 | No |
| 30 | Pdk3 | 18237 | -0.146 | -0.3951 | No |
| 31 | Lgals3 | 18736 | -0.161 | -0.4052 | No |
| 32 | Ech1 | 18884 | -0.167 | -0.4023 | No |
| 33 | Cyp51 | 19136 | -0.175 | -0.4028 | No |
| 34 | Actg1 | 19516 | -0.189 | -0.4072 | No |
| 35 | Mvd | 19541 | -0.190 | -0.3987 | No |
| 36 | Sc5d | 20063 | -0.208 | -0.4074 | No |
| 37 | Atf3 | 20504 | -0.220 | -0.4125 | No |
| 38 | Adh4 | 20695 | -0.227 | -0.4082 | No |
| 39 | Pcyt2 | 21384 | -0.251 | -0.4207 | Yes |
| 40 | Gusb | 21424 | -0.253 | -0.4096 | Yes |
| 41 | Fdft1 | 21920 | -0.267 | -0.4144 | Yes |
| 42 | Lss | 21981 | -0.270 | -0.4033 | Yes |
| 43 | Abca2 | 22132 | -0.277 | -0.3950 | Yes |
| 44 | Pparg | 22135 | -0.277 | -0.3814 | Yes |
| 45 | Aldoc | 22168 | -0.278 | -0.3688 | Yes |
| 46 | Tmem97 | 22435 | -0.290 | -0.3642 | Yes |
| 47 | Lgmn | 22807 | -0.301 | -0.3628 | Yes |
| 48 | Gstm7 | 23411 | -0.330 | -0.3684 | Yes |
| 49 | Hmgcs1 | 23491 | -0.334 | -0.3547 | Yes |
| 50 | Plaur | 23748 | -0.345 | -0.3470 | Yes |
| 51 | Pmvk | 23898 | -0.354 | -0.3349 | Yes |
| 52 | Acat2 | 23976 | -0.359 | -0.3200 | Yes |
| 53 | Nfil3 | 24005 | -0.360 | -0.3032 | Yes |
| 54 | Cd9 | 24044 | -0.362 | -0.2867 | Yes |
| 55 | S100a11 | 24416 | -0.386 | -0.2811 | Yes |
| 56 | Anxa5 | 25386 | -0.437 | -0.2947 | Yes |
| 57 | Plscr1 | 25474 | -0.444 | -0.2759 | Yes |
| 58 | Ldlr | 25701 | -0.464 | -0.2612 | Yes |
| 59 | Trib3 | 25830 | -0.471 | -0.2426 | Yes |
| 60 | Fabp5 | 26173 | -0.508 | -0.2299 | Yes |
| 61 | Tnfrsf12a | 26227 | -0.515 | -0.2064 | Yes |
| 62 | Alcam | 26333 | -0.529 | -0.1840 | Yes |
| 63 | Fdps | 26383 | -0.535 | -0.1594 | Yes |
| 64 | Hsd17b7 | 26787 | -0.603 | -0.1442 | Yes |
| 65 | Mvk | 26919 | -0.625 | -0.1181 | Yes |
| 66 | Niban1 | 27012 | -0.641 | -0.0898 | Yes |
| 67 | Gldc | 27099 | -0.667 | -0.0600 | Yes |
| 68 | Fbxo6 | 27261 | -0.719 | -0.0303 | Yes |
| 69 | Ebp | 27480 | -0.864 | 0.0044 | Yes |