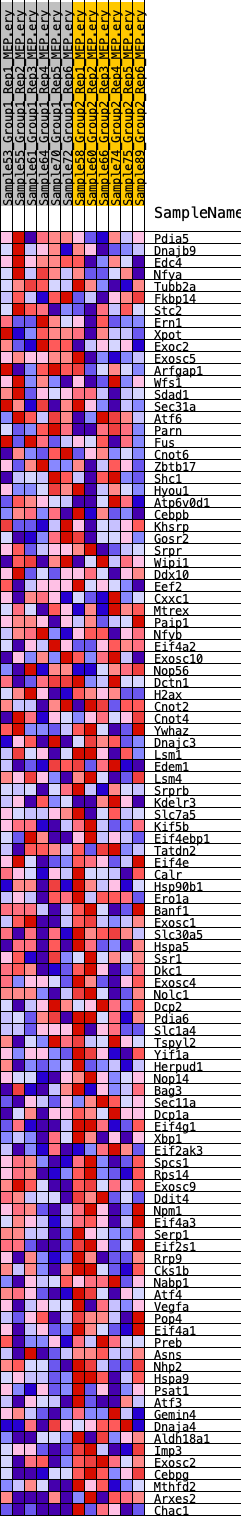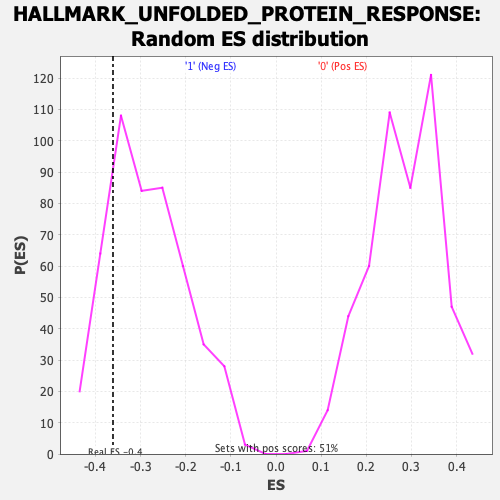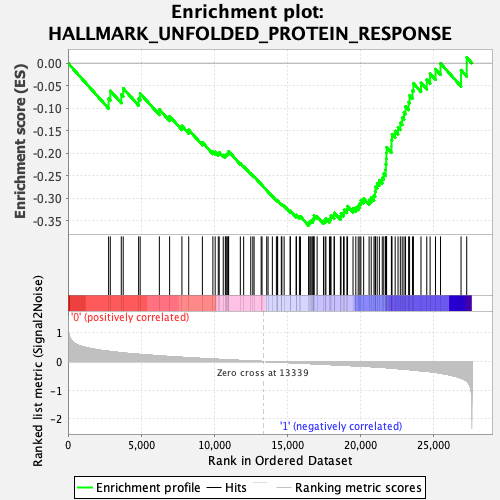
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group2.MEP.ery_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.36123502 |
| Normalized Enrichment Score (NES) | -1.2687011 |
| Nominal p-value | 0.19096509 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.811 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pdia5 | 2775 | 0.353 | -0.0789 | No |
| 2 | Dnajb9 | 2891 | 0.345 | -0.0615 | No |
| 3 | Edc4 | 3642 | 0.303 | -0.0698 | No |
| 4 | Nfya | 3774 | 0.297 | -0.0561 | No |
| 5 | Tubb2a | 4827 | 0.248 | -0.0788 | No |
| 6 | Fkbp14 | 4933 | 0.243 | -0.0675 | No |
| 7 | Stc2 | 6245 | 0.192 | -0.1031 | No |
| 8 | Ern1 | 6945 | 0.169 | -0.1180 | No |
| 9 | Xpot | 7787 | 0.143 | -0.1396 | No |
| 10 | Exoc2 | 8251 | 0.129 | -0.1484 | No |
| 11 | Exosc5 | 9186 | 0.102 | -0.1760 | No |
| 12 | Arfgap1 | 9903 | 0.083 | -0.1968 | No |
| 13 | Wfs1 | 10060 | 0.079 | -0.1975 | No |
| 14 | Sdad1 | 10272 | 0.073 | -0.2007 | No |
| 15 | Sec31a | 10335 | 0.071 | -0.1985 | No |
| 16 | Atf6 | 10611 | 0.064 | -0.2045 | No |
| 17 | Parn | 10755 | 0.061 | -0.2059 | No |
| 18 | Fus | 10801 | 0.059 | -0.2038 | No |
| 19 | Cnot6 | 10862 | 0.058 | -0.2024 | No |
| 20 | Zbtb17 | 10923 | 0.056 | -0.2010 | No |
| 21 | Shc1 | 10967 | 0.055 | -0.1992 | No |
| 22 | Hyou1 | 10980 | 0.055 | -0.1962 | No |
| 23 | Atp6v0d1 | 11779 | 0.034 | -0.2231 | No |
| 24 | Cebpb | 12007 | 0.031 | -0.2294 | No |
| 25 | Khsrp | 12492 | 0.019 | -0.2458 | No |
| 26 | Gosr2 | 12633 | 0.016 | -0.2499 | No |
| 27 | Srpr | 12720 | 0.015 | -0.2521 | No |
| 28 | Wipi1 | 13203 | 0.003 | -0.2694 | No |
| 29 | Ddx10 | 13262 | 0.002 | -0.2715 | No |
| 30 | Eef2 | 13583 | -0.002 | -0.2830 | No |
| 31 | Cxxc1 | 13675 | -0.004 | -0.2861 | No |
| 32 | Mtrex | 13969 | -0.010 | -0.2961 | No |
| 33 | Paip1 | 14244 | -0.015 | -0.3051 | No |
| 34 | Nfyb | 14310 | -0.016 | -0.3064 | No |
| 35 | Eif4a2 | 14330 | -0.017 | -0.3061 | No |
| 36 | Exosc10 | 14581 | -0.022 | -0.3138 | No |
| 37 | Nop56 | 14620 | -0.023 | -0.3137 | No |
| 38 | Dctn1 | 14773 | -0.027 | -0.3175 | No |
| 39 | H2ax | 15188 | -0.037 | -0.3303 | No |
| 40 | Cnot2 | 15193 | -0.037 | -0.3281 | No |
| 41 | Cnot4 | 15590 | -0.047 | -0.3396 | No |
| 42 | Ywhaz | 15617 | -0.047 | -0.3376 | No |
| 43 | Dnajc3 | 15819 | -0.051 | -0.3417 | No |
| 44 | Lsm1 | 15889 | -0.053 | -0.3409 | No |
| 45 | Edem1 | 16448 | -0.067 | -0.3571 | Yes |
| 46 | Lsm4 | 16477 | -0.067 | -0.3539 | Yes |
| 47 | Srprb | 16554 | -0.069 | -0.3523 | Yes |
| 48 | Kdelr3 | 16623 | -0.071 | -0.3503 | Yes |
| 49 | Slc7a5 | 16728 | -0.074 | -0.3495 | Yes |
| 50 | Kif5b | 16764 | -0.075 | -0.3461 | Yes |
| 51 | Eif4ebp1 | 16813 | -0.076 | -0.3431 | Yes |
| 52 | Tatdn2 | 16815 | -0.076 | -0.3384 | Yes |
| 53 | Eif4e | 17028 | -0.081 | -0.3410 | Yes |
| 54 | Calr | 17463 | -0.091 | -0.3511 | Yes |
| 55 | Hsp90b1 | 17575 | -0.094 | -0.3492 | Yes |
| 56 | Ero1a | 17629 | -0.096 | -0.3452 | Yes |
| 57 | Banf1 | 17871 | -0.102 | -0.3476 | Yes |
| 58 | Exosc1 | 17937 | -0.104 | -0.3435 | Yes |
| 59 | Slc30a5 | 17979 | -0.105 | -0.3384 | Yes |
| 60 | Hspa5 | 18189 | -0.109 | -0.3392 | Yes |
| 61 | Ssr1 | 18206 | -0.110 | -0.3329 | Yes |
| 62 | Dkc1 | 18620 | -0.120 | -0.3404 | Yes |
| 63 | Exosc4 | 18661 | -0.121 | -0.3343 | Yes |
| 64 | Nolc1 | 18834 | -0.126 | -0.3327 | Yes |
| 65 | Dcp2 | 18874 | -0.127 | -0.3262 | Yes |
| 66 | Pdia6 | 19065 | -0.132 | -0.3248 | Yes |
| 67 | Slc1a4 | 19108 | -0.133 | -0.3181 | Yes |
| 68 | Tspyl2 | 19483 | -0.140 | -0.3229 | Yes |
| 69 | Yif1a | 19673 | -0.146 | -0.3207 | Yes |
| 70 | Herpud1 | 19837 | -0.149 | -0.3173 | Yes |
| 71 | Nop14 | 19949 | -0.153 | -0.3118 | Yes |
| 72 | Bag3 | 20020 | -0.155 | -0.3046 | Yes |
| 73 | Sec11a | 20209 | -0.161 | -0.3014 | Yes |
| 74 | Dcp1a | 20581 | -0.171 | -0.3042 | Yes |
| 75 | Eif4g1 | 20726 | -0.175 | -0.2986 | Yes |
| 76 | Xbp1 | 20915 | -0.182 | -0.2941 | Yes |
| 77 | Eif2ak3 | 20995 | -0.184 | -0.2854 | Yes |
| 78 | Spcs1 | 21020 | -0.185 | -0.2747 | Yes |
| 79 | Rps14 | 21133 | -0.188 | -0.2671 | Yes |
| 80 | Exosc9 | 21282 | -0.194 | -0.2603 | Yes |
| 81 | Ddit4 | 21482 | -0.201 | -0.2550 | Yes |
| 82 | Npm1 | 21585 | -0.206 | -0.2459 | Yes |
| 83 | Eif4a3 | 21710 | -0.209 | -0.2374 | Yes |
| 84 | Serp1 | 21723 | -0.209 | -0.2247 | Yes |
| 85 | Eif2s1 | 21754 | -0.211 | -0.2126 | Yes |
| 86 | Rrp9 | 21764 | -0.211 | -0.1998 | Yes |
| 87 | Cks1b | 21779 | -0.212 | -0.1870 | Yes |
| 88 | Nabp1 | 22108 | -0.225 | -0.1849 | Yes |
| 89 | Atf4 | 22109 | -0.225 | -0.1708 | Yes |
| 90 | Vegfa | 22147 | -0.228 | -0.1579 | Yes |
| 91 | Pop4 | 22363 | -0.236 | -0.1510 | Yes |
| 92 | Eif4a1 | 22559 | -0.245 | -0.1428 | Yes |
| 93 | Preb | 22715 | -0.251 | -0.1327 | Yes |
| 94 | Asns | 22843 | -0.257 | -0.1213 | Yes |
| 95 | Nhp2 | 22967 | -0.262 | -0.1094 | Yes |
| 96 | Hspa9 | 23071 | -0.268 | -0.0964 | Yes |
| 97 | Psat1 | 23284 | -0.277 | -0.0868 | Yes |
| 98 | Atf3 | 23348 | -0.278 | -0.0718 | Yes |
| 99 | Gemin4 | 23546 | -0.287 | -0.0610 | Yes |
| 100 | Dnaja4 | 23612 | -0.291 | -0.0452 | Yes |
| 101 | Aldh18a1 | 24128 | -0.319 | -0.0440 | Yes |
| 102 | Imp3 | 24523 | -0.340 | -0.0371 | Yes |
| 103 | Exosc2 | 24751 | -0.351 | -0.0235 | Yes |
| 104 | Cebpg | 25121 | -0.376 | -0.0134 | Yes |
| 105 | Mthfd2 | 25463 | -0.403 | -0.0006 | Yes |
| 106 | Arxes2 | 26866 | -0.576 | -0.0157 | Yes |
| 107 | Chac1 | 27254 | -0.679 | 0.0127 | Yes |