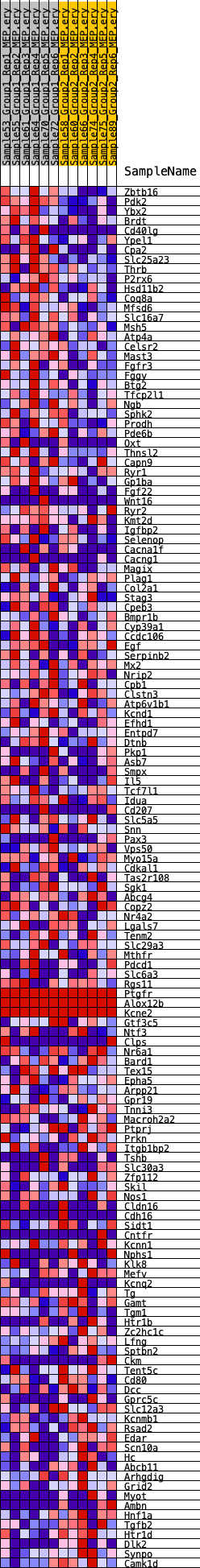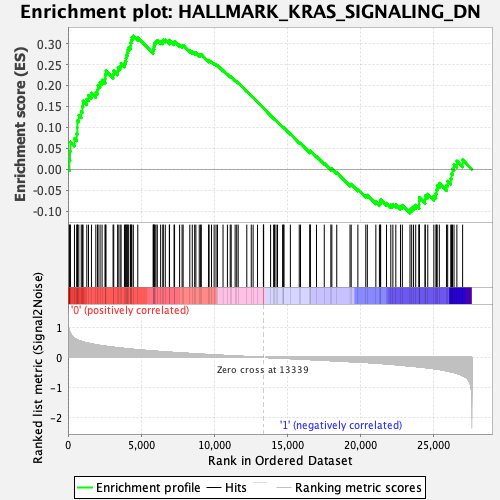
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group2.MEP.ery_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.3182968 |
| Normalized Enrichment Score (NES) | 1.2939762 |
| Nominal p-value | 0.018907564 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.819 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zbtb16 | 95 | 0.873 | 0.0216 | Yes |
| 2 | Pdk2 | 134 | 0.813 | 0.0436 | Yes |
| 3 | Ybx2 | 164 | 0.791 | 0.0652 | Yes |
| 4 | Brdt | 441 | 0.634 | 0.0734 | Yes |
| 5 | Cd40lg | 597 | 0.588 | 0.0846 | Yes |
| 6 | Ypel1 | 640 | 0.578 | 0.0997 | Yes |
| 7 | Cpa2 | 642 | 0.578 | 0.1162 | Yes |
| 8 | Slc25a23 | 737 | 0.559 | 0.1289 | Yes |
| 9 | Thrb | 905 | 0.529 | 0.1380 | Yes |
| 10 | P2rx6 | 981 | 0.516 | 0.1501 | Yes |
| 11 | Hsd11b2 | 1027 | 0.509 | 0.1631 | Yes |
| 12 | Coq8a | 1285 | 0.478 | 0.1674 | Yes |
| 13 | Mfsd6 | 1400 | 0.465 | 0.1766 | Yes |
| 14 | Slc16a7 | 1603 | 0.443 | 0.1820 | Yes |
| 15 | Msh5 | 1902 | 0.416 | 0.1830 | Yes |
| 16 | Atp4a | 2023 | 0.405 | 0.1903 | Yes |
| 17 | Celsr2 | 2050 | 0.403 | 0.2009 | Yes |
| 18 | Mast3 | 2191 | 0.392 | 0.2071 | Yes |
| 19 | Fgfr3 | 2329 | 0.382 | 0.2131 | Yes |
| 20 | Fggy | 2539 | 0.368 | 0.2160 | Yes |
| 21 | Btg2 | 2547 | 0.368 | 0.2263 | Yes |
| 22 | Tfcp2l1 | 2597 | 0.364 | 0.2350 | Yes |
| 23 | Ngb | 3082 | 0.334 | 0.2270 | Yes |
| 24 | Sphk2 | 3126 | 0.331 | 0.2349 | Yes |
| 25 | Prodh | 3398 | 0.317 | 0.2341 | Yes |
| 26 | Pde6b | 3423 | 0.315 | 0.2423 | Yes |
| 27 | Oxt | 3557 | 0.307 | 0.2463 | Yes |
| 28 | Thnsl2 | 3627 | 0.303 | 0.2525 | Yes |
| 29 | Capn9 | 3858 | 0.293 | 0.2525 | Yes |
| 30 | Ryr1 | 3919 | 0.290 | 0.2587 | Yes |
| 31 | Gp1ba | 3965 | 0.287 | 0.2652 | Yes |
| 32 | Fgf22 | 3983 | 0.285 | 0.2728 | Yes |
| 33 | Wnt16 | 4051 | 0.281 | 0.2785 | Yes |
| 34 | Ryr2 | 4096 | 0.280 | 0.2849 | Yes |
| 35 | Kmt2d | 4150 | 0.278 | 0.2910 | Yes |
| 36 | Igfbp2 | 4278 | 0.272 | 0.2942 | Yes |
| 37 | Selenop | 4283 | 0.272 | 0.3018 | Yes |
| 38 | Cacna1f | 4325 | 0.270 | 0.3081 | Yes |
| 39 | Cacng1 | 4362 | 0.267 | 0.3144 | Yes |
| 40 | Magix | 4464 | 0.263 | 0.3183 | Yes |
| 41 | Plag1 | 4770 | 0.250 | 0.3144 | No |
| 42 | Col2a1 | 5825 | 0.210 | 0.2820 | No |
| 43 | Stag3 | 5836 | 0.210 | 0.2877 | No |
| 44 | Cpeb3 | 5877 | 0.208 | 0.2922 | No |
| 45 | Bmpr1b | 5896 | 0.207 | 0.2975 | No |
| 46 | Cyp39a1 | 5939 | 0.205 | 0.3019 | No |
| 47 | Ccdc106 | 6011 | 0.202 | 0.3051 | No |
| 48 | Egf | 6103 | 0.198 | 0.3074 | No |
| 49 | Serpinb2 | 6332 | 0.190 | 0.3046 | No |
| 50 | Mx2 | 6485 | 0.185 | 0.3044 | No |
| 51 | Nrip2 | 6508 | 0.184 | 0.3088 | No |
| 52 | Cpb1 | 6649 | 0.179 | 0.3089 | No |
| 53 | Clstn3 | 6927 | 0.170 | 0.3037 | No |
| 54 | Atp6v1b1 | 6940 | 0.170 | 0.3081 | No |
| 55 | Kcnd1 | 7250 | 0.159 | 0.3014 | No |
| 56 | Efhd1 | 7283 | 0.157 | 0.3048 | No |
| 57 | Entpd7 | 7627 | 0.148 | 0.2965 | No |
| 58 | Dtnb | 7797 | 0.143 | 0.2945 | No |
| 59 | Pkp1 | 7870 | 0.140 | 0.2959 | No |
| 60 | Asb7 | 8325 | 0.127 | 0.2830 | No |
| 61 | Smpx | 8495 | 0.122 | 0.2804 | No |
| 62 | Il5 | 8658 | 0.117 | 0.2778 | No |
| 63 | Tcf7l1 | 8741 | 0.115 | 0.2782 | No |
| 64 | Idua | 8971 | 0.108 | 0.2729 | No |
| 65 | Cd207 | 9065 | 0.105 | 0.2726 | No |
| 66 | Slc5a5 | 9111 | 0.104 | 0.2739 | No |
| 67 | Snn | 9608 | 0.091 | 0.2585 | No |
| 68 | Pax3 | 9643 | 0.090 | 0.2598 | No |
| 69 | Vps50 | 9808 | 0.085 | 0.2563 | No |
| 70 | Myo15a | 9988 | 0.081 | 0.2521 | No |
| 71 | Cdkal1 | 10119 | 0.077 | 0.2496 | No |
| 72 | Tas2r108 | 10216 | 0.075 | 0.2482 | No |
| 73 | Sgk1 | 10597 | 0.065 | 0.2363 | No |
| 74 | Abcg4 | 10896 | 0.057 | 0.2270 | No |
| 75 | Copz2 | 11087 | 0.052 | 0.2216 | No |
| 76 | Nr4a2 | 11154 | 0.050 | 0.2206 | No |
| 77 | Lgals7 | 11424 | 0.044 | 0.2121 | No |
| 78 | Tenm2 | 11533 | 0.041 | 0.2093 | No |
| 79 | Slc29a3 | 11632 | 0.038 | 0.2069 | No |
| 80 | Mthfr | 12221 | 0.025 | 0.1862 | No |
| 81 | Pdcd1 | 12526 | 0.019 | 0.1756 | No |
| 82 | Slc6a3 | 12654 | 0.016 | 0.1715 | No |
| 83 | Rgs11 | 12952 | 0.009 | 0.1609 | No |
| 84 | Ptgfr | 13355 | 0.000 | 0.1463 | No |
| 85 | Alox12b | 13365 | 0.000 | 0.1459 | No |
| 86 | Kcne2 | 13372 | 0.000 | 0.1457 | No |
| 87 | Gtf3c5 | 13848 | -0.007 | 0.1286 | No |
| 88 | Ntf3 | 14058 | -0.012 | 0.1213 | No |
| 89 | Clps | 14117 | -0.013 | 0.1196 | No |
| 90 | Nr6a1 | 14219 | -0.015 | 0.1164 | No |
| 91 | Bard1 | 14322 | -0.017 | 0.1131 | No |
| 92 | Tex15 | 14668 | -0.024 | 0.1013 | No |
| 93 | Epha5 | 14740 | -0.026 | 0.0994 | No |
| 94 | Arpp21 | 14761 | -0.027 | 0.0995 | No |
| 95 | Gpr19 | 15200 | -0.037 | 0.0846 | No |
| 96 | Tnni3 | 15807 | -0.051 | 0.0640 | No |
| 97 | Macroh2a2 | 15893 | -0.053 | 0.0624 | No |
| 98 | Ptprj | 16539 | -0.069 | 0.0409 | No |
| 99 | Prkn | 16544 | -0.069 | 0.0427 | No |
| 100 | Itgb1bp2 | 16556 | -0.069 | 0.0443 | No |
| 101 | Tshb | 16986 | -0.080 | 0.0310 | No |
| 102 | Slc30a3 | 17517 | -0.093 | 0.0144 | No |
| 103 | Zfp112 | 17967 | -0.105 | 0.0010 | No |
| 104 | Skil | 18035 | -0.106 | 0.0016 | No |
| 105 | Nos1 | 18370 | -0.113 | -0.0073 | No |
| 106 | Cldn16 | 19277 | -0.138 | -0.0363 | No |
| 107 | Cdh16 | 19358 | -0.140 | -0.0352 | No |
| 108 | Sidt1 | 19814 | -0.148 | -0.0475 | No |
| 109 | Cntfr | 20354 | -0.165 | -0.0624 | No |
| 110 | Kcnn1 | 20461 | -0.167 | -0.0615 | No |
| 111 | Nphs1 | 21042 | -0.185 | -0.0773 | No |
| 112 | Klk8 | 21286 | -0.194 | -0.0806 | No |
| 113 | Mefv | 21348 | -0.196 | -0.0771 | No |
| 114 | Kcnq2 | 21371 | -0.197 | -0.0723 | No |
| 115 | Tg | 21770 | -0.212 | -0.0807 | No |
| 116 | Gamt | 22064 | -0.223 | -0.0850 | No |
| 117 | Tgm1 | 22207 | -0.231 | -0.0835 | No |
| 118 | Htr1b | 22408 | -0.238 | -0.0839 | No |
| 119 | Zc2hc1c | 22727 | -0.252 | -0.0883 | No |
| 120 | Lfng | 22854 | -0.258 | -0.0855 | No |
| 121 | Sptbn2 | 23377 | -0.280 | -0.0965 | No |
| 122 | Ckm | 23492 | -0.286 | -0.0924 | No |
| 123 | Tent5c | 23620 | -0.292 | -0.0886 | No |
| 124 | Cd80 | 23767 | -0.299 | -0.0854 | No |
| 125 | Dcc | 23998 | -0.314 | -0.0847 | No |
| 126 | Gprc5c | 24002 | -0.315 | -0.0758 | No |
| 127 | Slc12a3 | 24007 | -0.315 | -0.0669 | No |
| 128 | Kcnmb1 | 24403 | -0.332 | -0.0717 | No |
| 129 | Rsad2 | 24421 | -0.334 | -0.0628 | No |
| 130 | Edar | 24591 | -0.342 | -0.0591 | No |
| 131 | Scn10a | 25007 | -0.369 | -0.0636 | No |
| 132 | Hc | 25150 | -0.378 | -0.0580 | No |
| 133 | Abcb11 | 25193 | -0.380 | -0.0486 | No |
| 134 | Arhgdig | 25234 | -0.385 | -0.0390 | No |
| 135 | Grid2 | 25381 | -0.397 | -0.0329 | No |
| 136 | Myot | 25875 | -0.441 | -0.0382 | No |
| 137 | Ambn | 25952 | -0.447 | -0.0281 | No |
| 138 | Hnf1a | 26164 | -0.474 | -0.0222 | No |
| 139 | Tgfb2 | 26215 | -0.479 | -0.0103 | No |
| 140 | Htr1d | 26284 | -0.488 | 0.0013 | No |
| 141 | Dlk2 | 26386 | -0.498 | 0.0119 | No |
| 142 | Synpo | 26581 | -0.524 | 0.0199 | No |
| 143 | Camk1d | 26974 | -0.601 | 0.0229 | No |