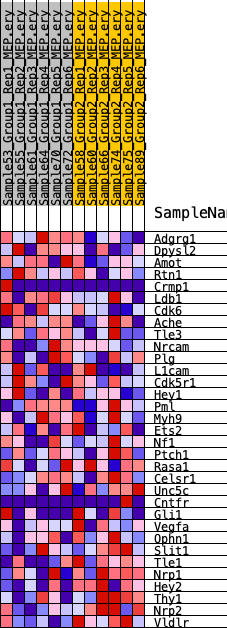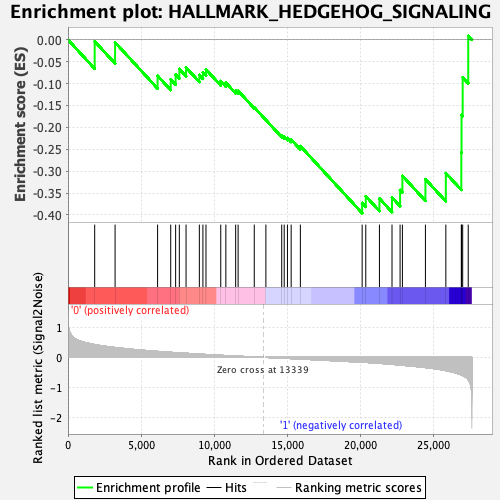
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group2.MEP.ery_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | -0.39597255 |
| Normalized Enrichment Score (NES) | -1.1860806 |
| Nominal p-value | 0.2718053 |
| FDR q-value | 0.72863805 |
| FWER p-Value | 0.898 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adgrg1 | 1824 | 0.423 | -0.0037 | No |
| 2 | Dpysl2 | 3218 | 0.326 | -0.0061 | No |
| 3 | Amot | 6124 | 0.197 | -0.0824 | No |
| 4 | Rtn1 | 7021 | 0.166 | -0.0903 | No |
| 5 | Crmp1 | 7357 | 0.155 | -0.0796 | No |
| 6 | Ldb1 | 7609 | 0.149 | -0.0667 | No |
| 7 | Cdk6 | 8069 | 0.135 | -0.0634 | No |
| 8 | Ache | 8978 | 0.108 | -0.0804 | No |
| 9 | Tle3 | 9222 | 0.101 | -0.0743 | No |
| 10 | Nrcam | 9431 | 0.095 | -0.0678 | No |
| 11 | Plg | 10441 | 0.068 | -0.0944 | No |
| 12 | L1cam | 10790 | 0.060 | -0.0982 | No |
| 13 | Cdk5r1 | 11460 | 0.043 | -0.1162 | No |
| 14 | Hey1 | 11628 | 0.038 | -0.1166 | No |
| 15 | Pml | 12732 | 0.014 | -0.1545 | No |
| 16 | Myh9 | 13526 | -0.000 | -0.1832 | No |
| 17 | Ets2 | 14608 | -0.023 | -0.2191 | No |
| 18 | Nf1 | 14778 | -0.028 | -0.2211 | No |
| 19 | Ptch1 | 15000 | -0.033 | -0.2243 | No |
| 20 | Rasa1 | 15254 | -0.038 | -0.2278 | No |
| 21 | Celsr1 | 15883 | -0.053 | -0.2428 | No |
| 22 | Unc5c | 20106 | -0.158 | -0.3727 | Yes |
| 23 | Cntfr | 20354 | -0.165 | -0.3573 | Yes |
| 24 | Gli1 | 21288 | -0.194 | -0.3625 | Yes |
| 25 | Vegfa | 22147 | -0.228 | -0.3600 | Yes |
| 26 | Ophn1 | 22699 | -0.250 | -0.3431 | Yes |
| 27 | Slit1 | 22858 | -0.258 | -0.3108 | Yes |
| 28 | Tle1 | 24431 | -0.334 | -0.3185 | Yes |
| 29 | Nrp1 | 25827 | -0.437 | -0.3046 | Yes |
| 30 | Hey2 | 26884 | -0.580 | -0.2574 | Yes |
| 31 | Thy1 | 26900 | -0.583 | -0.1718 | Yes |
| 32 | Nrp2 | 26978 | -0.602 | -0.0859 | Yes |
| 33 | Vldlr | 27357 | -0.735 | 0.0089 | Yes |