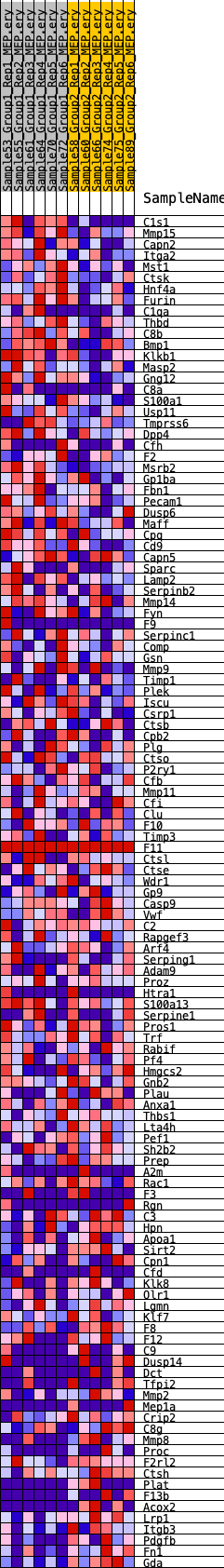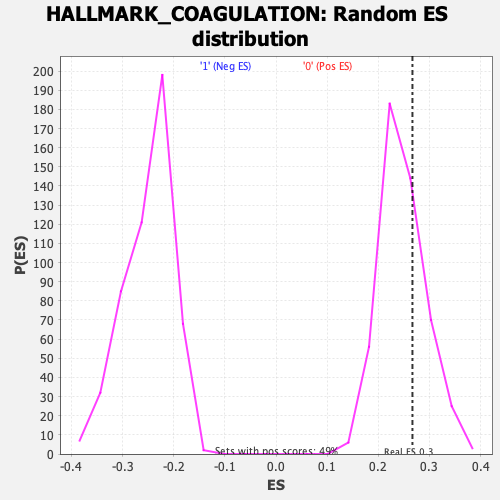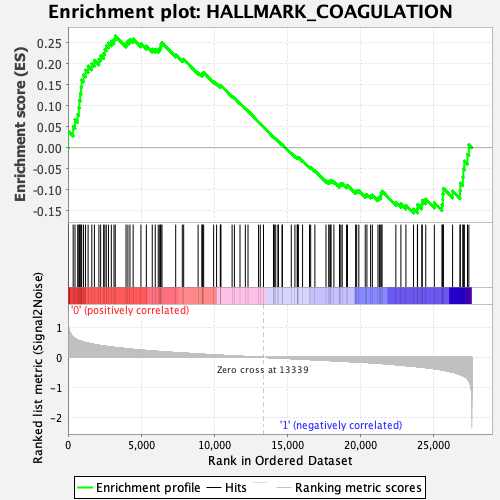
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group2.MEP.ery_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.266778 |
| Normalized Enrichment Score (NES) | 1.0772704 |
| Nominal p-value | 0.27926078 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.96 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | C1s1 | 10 | 1.208 | 0.0405 | Yes |
| 2 | Mmp15 | 353 | 0.668 | 0.0506 | Yes |
| 3 | Capn2 | 478 | 0.620 | 0.0671 | Yes |
| 4 | Itga2 | 662 | 0.575 | 0.0799 | Yes |
| 5 | Mst1 | 738 | 0.559 | 0.0960 | Yes |
| 6 | Ctsk | 780 | 0.550 | 0.1131 | Yes |
| 7 | Hnf4a | 837 | 0.540 | 0.1294 | Yes |
| 8 | Furin | 894 | 0.531 | 0.1453 | Yes |
| 9 | C1qa | 935 | 0.522 | 0.1615 | Yes |
| 10 | Thbd | 1062 | 0.503 | 0.1739 | Yes |
| 11 | C8b | 1200 | 0.488 | 0.1854 | Yes |
| 12 | Bmp1 | 1374 | 0.468 | 0.1949 | Yes |
| 13 | Klkb1 | 1628 | 0.440 | 0.2006 | Yes |
| 14 | Masp2 | 1805 | 0.425 | 0.2086 | Yes |
| 15 | Gng12 | 2113 | 0.398 | 0.2109 | Yes |
| 16 | C8a | 2232 | 0.388 | 0.2197 | Yes |
| 17 | S100a1 | 2435 | 0.375 | 0.2250 | Yes |
| 18 | Usp11 | 2518 | 0.369 | 0.2345 | Yes |
| 19 | Tmprss6 | 2615 | 0.364 | 0.2433 | Yes |
| 20 | Dpp4 | 2765 | 0.354 | 0.2499 | Yes |
| 21 | Cfh | 2961 | 0.340 | 0.2543 | Yes |
| 22 | F2 | 3144 | 0.330 | 0.2588 | Yes |
| 23 | Msrb2 | 3229 | 0.326 | 0.2668 | Yes |
| 24 | Gp1ba | 3965 | 0.287 | 0.2497 | No |
| 25 | Fbn1 | 4095 | 0.280 | 0.2545 | No |
| 26 | Pecam1 | 4234 | 0.274 | 0.2588 | No |
| 27 | Dusp6 | 4454 | 0.263 | 0.2597 | No |
| 28 | Maff | 4986 | 0.241 | 0.2485 | No |
| 29 | Cpq | 5355 | 0.227 | 0.2428 | No |
| 30 | Cd9 | 5759 | 0.212 | 0.2353 | No |
| 31 | Capn5 | 5963 | 0.204 | 0.2348 | No |
| 32 | Sparc | 6169 | 0.196 | 0.2340 | No |
| 33 | Lamp2 | 6269 | 0.192 | 0.2369 | No |
| 34 | Serpinb2 | 6332 | 0.190 | 0.2410 | No |
| 35 | Mmp14 | 6347 | 0.189 | 0.2469 | No |
| 36 | Fyn | 6424 | 0.187 | 0.2505 | No |
| 37 | F9 | 7361 | 0.155 | 0.2217 | No |
| 38 | Serpinc1 | 7812 | 0.142 | 0.2101 | No |
| 39 | Comp | 7907 | 0.139 | 0.2114 | No |
| 40 | Gsn | 8894 | 0.111 | 0.1793 | No |
| 41 | Mmp9 | 9159 | 0.103 | 0.1731 | No |
| 42 | Timp1 | 9167 | 0.103 | 0.1763 | No |
| 43 | Plek | 9213 | 0.101 | 0.1781 | No |
| 44 | Iscu | 9274 | 0.100 | 0.1793 | No |
| 45 | Csrp1 | 9955 | 0.082 | 0.1573 | No |
| 46 | Ctsb | 10156 | 0.076 | 0.1527 | No |
| 47 | Cpb2 | 10409 | 0.069 | 0.1458 | No |
| 48 | Plg | 10441 | 0.068 | 0.1470 | No |
| 49 | Ctso | 10442 | 0.068 | 0.1493 | No |
| 50 | P2ry1 | 11217 | 0.048 | 0.1228 | No |
| 51 | Cfb | 11373 | 0.045 | 0.1186 | No |
| 52 | Mmp11 | 11757 | 0.035 | 0.1059 | No |
| 53 | Cfi | 12122 | 0.028 | 0.0936 | No |
| 54 | Clu | 12304 | 0.023 | 0.0878 | No |
| 55 | F10 | 13025 | 0.007 | 0.0618 | No |
| 56 | Timp3 | 13142 | 0.004 | 0.0578 | No |
| 57 | F11 | 13363 | 0.000 | 0.0498 | No |
| 58 | Ctsl | 14045 | -0.012 | 0.0254 | No |
| 59 | Ctse | 14079 | -0.012 | 0.0246 | No |
| 60 | Wdr1 | 14092 | -0.013 | 0.0246 | No |
| 61 | Gp9 | 14198 | -0.014 | 0.0212 | No |
| 62 | Casp9 | 14350 | -0.017 | 0.0163 | No |
| 63 | Vwf | 14382 | -0.018 | 0.0158 | No |
| 64 | C2 | 14630 | -0.023 | 0.0076 | No |
| 65 | Rapgef3 | 14655 | -0.024 | 0.0076 | No |
| 66 | Arf4 | 15261 | -0.038 | -0.0131 | No |
| 67 | Serping1 | 15511 | -0.044 | -0.0207 | No |
| 68 | Adam9 | 15664 | -0.048 | -0.0246 | No |
| 69 | Proz | 15701 | -0.049 | -0.0243 | No |
| 70 | Htra1 | 15710 | -0.049 | -0.0229 | No |
| 71 | S100a13 | 15743 | -0.050 | -0.0224 | No |
| 72 | Serpine1 | 16034 | -0.056 | -0.0310 | No |
| 73 | Pros1 | 16508 | -0.068 | -0.0459 | No |
| 74 | Trf | 16588 | -0.070 | -0.0464 | No |
| 75 | Rabif | 16876 | -0.077 | -0.0543 | No |
| 76 | Pf4 | 17635 | -0.096 | -0.0786 | No |
| 77 | Hmgcs2 | 17831 | -0.101 | -0.0823 | No |
| 78 | Gnb2 | 17846 | -0.101 | -0.0793 | No |
| 79 | Plau | 17955 | -0.104 | -0.0797 | No |
| 80 | Anxa1 | 17975 | -0.105 | -0.0769 | No |
| 81 | Thbs1 | 18176 | -0.109 | -0.0805 | No |
| 82 | Lta4h | 18557 | -0.118 | -0.0903 | No |
| 83 | Pef1 | 18575 | -0.118 | -0.0869 | No |
| 84 | Sh2b2 | 18652 | -0.121 | -0.0856 | No |
| 85 | Prep | 18744 | -0.123 | -0.0848 | No |
| 86 | A2m | 19041 | -0.132 | -0.0911 | No |
| 87 | Rac1 | 19103 | -0.133 | -0.0888 | No |
| 88 | F3 | 19660 | -0.145 | -0.1041 | No |
| 89 | Rgn | 19731 | -0.147 | -0.1017 | No |
| 90 | C3 | 19879 | -0.150 | -0.1020 | No |
| 91 | Hpn | 20317 | -0.164 | -0.1123 | No |
| 92 | Apoa1 | 20432 | -0.166 | -0.1108 | No |
| 93 | Sirt2 | 20684 | -0.174 | -0.1141 | No |
| 94 | Cpn1 | 20803 | -0.178 | -0.1124 | No |
| 95 | Cfd | 21188 | -0.191 | -0.1199 | No |
| 96 | Klk8 | 21286 | -0.194 | -0.1169 | No |
| 97 | Olr1 | 21388 | -0.198 | -0.1139 | No |
| 98 | Lgmn | 21396 | -0.198 | -0.1074 | No |
| 99 | Klf7 | 21468 | -0.200 | -0.1032 | No |
| 100 | F8 | 22411 | -0.238 | -0.1295 | No |
| 101 | F12 | 22755 | -0.253 | -0.1334 | No |
| 102 | C9 | 23098 | -0.270 | -0.1367 | No |
| 103 | Dusp14 | 23618 | -0.291 | -0.1458 | No |
| 104 | Dct | 23882 | -0.307 | -0.1449 | No |
| 105 | Tfpi2 | 23893 | -0.308 | -0.1349 | No |
| 106 | Mmp2 | 24185 | -0.322 | -0.1346 | No |
| 107 | Mep1a | 24212 | -0.324 | -0.1246 | No |
| 108 | Crip2 | 24454 | -0.336 | -0.1220 | No |
| 109 | C8g | 25041 | -0.372 | -0.1308 | No |
| 110 | Mmp8 | 25562 | -0.413 | -0.1357 | No |
| 111 | Proc | 25619 | -0.418 | -0.1237 | No |
| 112 | F2rl2 | 25624 | -0.418 | -0.1097 | No |
| 113 | Ctsh | 25658 | -0.421 | -0.0966 | No |
| 114 | Plat | 26289 | -0.488 | -0.1030 | No |
| 115 | F13b | 26795 | -0.564 | -0.1023 | No |
| 116 | Acox2 | 26813 | -0.567 | -0.0838 | No |
| 117 | Lrp1 | 26988 | -0.604 | -0.0697 | No |
| 118 | Itgb3 | 27025 | -0.612 | -0.0503 | No |
| 119 | Pdgfb | 27093 | -0.631 | -0.0314 | No |
| 120 | Fn1 | 27307 | -0.705 | -0.0153 | No |
| 121 | Gda | 27401 | -0.769 | 0.0073 | No |