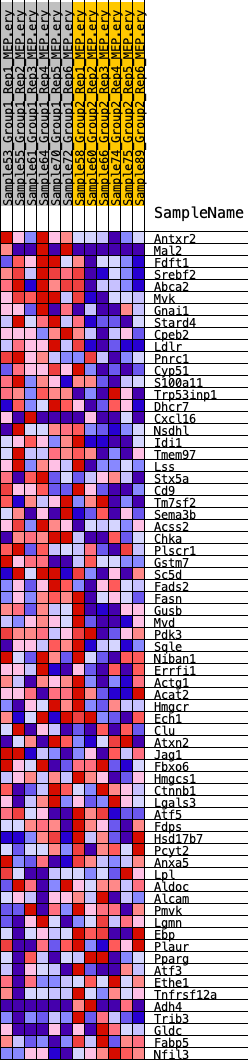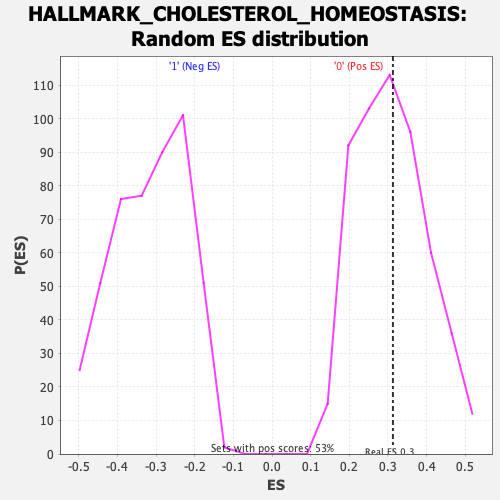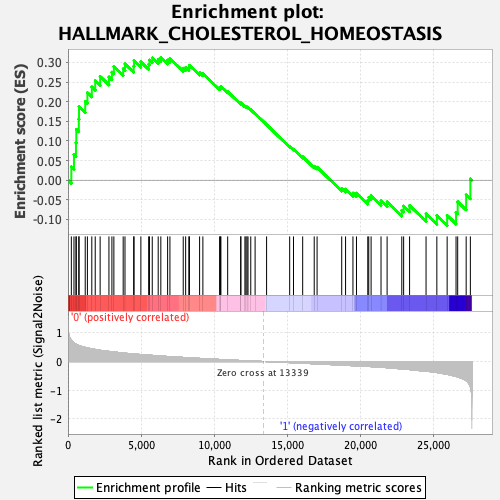
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group2.MEP.ery_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.31290042 |
| Normalized Enrichment Score (NES) | 1.0111794 |
| Nominal p-value | 0.47817838 |
| FDR q-value | 0.81254613 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Antxr2 | 228 | 0.732 | 0.0344 | Yes |
| 2 | Mal2 | 408 | 0.644 | 0.0654 | Yes |
| 3 | Fdft1 | 544 | 0.602 | 0.0955 | Yes |
| 4 | Srebf2 | 567 | 0.596 | 0.1294 | Yes |
| 5 | Abca2 | 740 | 0.559 | 0.1557 | Yes |
| 6 | Mvk | 755 | 0.555 | 0.1876 | Yes |
| 7 | Gnai1 | 1177 | 0.491 | 0.2009 | Yes |
| 8 | Stard4 | 1326 | 0.473 | 0.2231 | Yes |
| 9 | Cpeb2 | 1627 | 0.440 | 0.2378 | Yes |
| 10 | Ldlr | 1861 | 0.420 | 0.2538 | Yes |
| 11 | Pnrc1 | 2196 | 0.391 | 0.2645 | Yes |
| 12 | Cyp51 | 2798 | 0.352 | 0.2631 | Yes |
| 13 | S100a11 | 3001 | 0.338 | 0.2754 | Yes |
| 14 | Trp53inp1 | 3135 | 0.331 | 0.2899 | Yes |
| 15 | Dhcr7 | 3772 | 0.297 | 0.2840 | Yes |
| 16 | Cxcl16 | 3895 | 0.291 | 0.2965 | Yes |
| 17 | Nsdhl | 4489 | 0.262 | 0.2903 | Yes |
| 18 | Idi1 | 4515 | 0.261 | 0.3046 | Yes |
| 19 | Tmem97 | 4981 | 0.241 | 0.3017 | Yes |
| 20 | Lss | 5514 | 0.222 | 0.2953 | Yes |
| 21 | Stx5a | 5568 | 0.220 | 0.3062 | Yes |
| 22 | Cd9 | 5759 | 0.212 | 0.3117 | Yes |
| 23 | Tm7sf2 | 6165 | 0.196 | 0.3084 | Yes |
| 24 | Sema3b | 6345 | 0.189 | 0.3129 | Yes |
| 25 | Acss2 | 6809 | 0.173 | 0.3062 | No |
| 26 | Chka | 6967 | 0.169 | 0.3103 | No |
| 27 | Plscr1 | 7876 | 0.140 | 0.2855 | No |
| 28 | Gstm7 | 8042 | 0.136 | 0.2874 | No |
| 29 | Sc5d | 8261 | 0.129 | 0.2870 | No |
| 30 | Fads2 | 8301 | 0.128 | 0.2930 | No |
| 31 | Fasn | 8995 | 0.108 | 0.2741 | No |
| 32 | Gusb | 9219 | 0.101 | 0.2719 | No |
| 33 | Mvd | 10362 | 0.070 | 0.2345 | No |
| 34 | Pdk3 | 10420 | 0.069 | 0.2364 | No |
| 35 | Sqle | 10451 | 0.068 | 0.2393 | No |
| 36 | Niban1 | 10914 | 0.056 | 0.2258 | No |
| 37 | Errfi1 | 11809 | 0.034 | 0.1953 | No |
| 38 | Actg1 | 11812 | 0.034 | 0.1972 | No |
| 39 | Acat2 | 12093 | 0.028 | 0.1887 | No |
| 40 | Hmgcr | 12162 | 0.027 | 0.1878 | No |
| 41 | Ech1 | 12236 | 0.025 | 0.1866 | No |
| 42 | Clu | 12304 | 0.023 | 0.1855 | No |
| 43 | Atxn2 | 12493 | 0.019 | 0.1798 | No |
| 44 | Jag1 | 12789 | 0.013 | 0.1699 | No |
| 45 | Fbxo6 | 13574 | -0.001 | 0.1415 | No |
| 46 | Hmgcs1 | 15152 | -0.036 | 0.0863 | No |
| 47 | Ctnnb1 | 15413 | -0.042 | 0.0793 | No |
| 48 | Lgals3 | 16041 | -0.057 | 0.0598 | No |
| 49 | Atf5 | 16830 | -0.076 | 0.0356 | No |
| 50 | Fdps | 17026 | -0.081 | 0.0333 | No |
| 51 | Hsd17b7 | 18709 | -0.122 | -0.0207 | No |
| 52 | Pcyt2 | 18973 | -0.130 | -0.0227 | No |
| 53 | Anxa5 | 19477 | -0.140 | -0.0328 | No |
| 54 | Lpl | 19718 | -0.147 | -0.0329 | No |
| 55 | Aldoc | 20491 | -0.168 | -0.0512 | No |
| 56 | Alcam | 20557 | -0.170 | -0.0437 | No |
| 57 | Pmvk | 20718 | -0.175 | -0.0393 | No |
| 58 | Lgmn | 21396 | -0.198 | -0.0524 | No |
| 59 | Ebp | 21812 | -0.213 | -0.0550 | No |
| 60 | Plaur | 22816 | -0.256 | -0.0765 | No |
| 61 | Pparg | 22942 | -0.261 | -0.0659 | No |
| 62 | Atf3 | 23348 | -0.278 | -0.0644 | No |
| 63 | Ethe1 | 24474 | -0.336 | -0.0857 | No |
| 64 | Tnfrsf12a | 25212 | -0.382 | -0.0902 | No |
| 65 | Adh4 | 25916 | -0.444 | -0.0898 | No |
| 66 | Trib3 | 26524 | -0.515 | -0.0819 | No |
| 67 | Gldc | 26636 | -0.536 | -0.0547 | No |
| 68 | Fabp5 | 27216 | -0.663 | -0.0371 | No |
| 69 | Nfil3 | 27503 | -0.877 | 0.0036 | No |