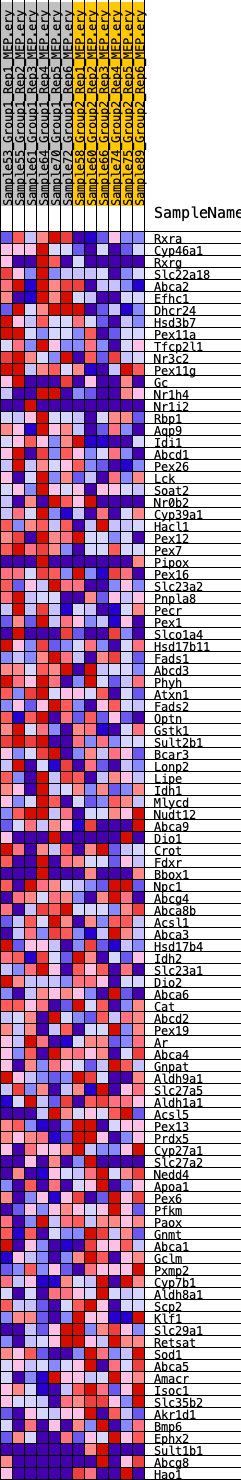
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group2.MEP.ery_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
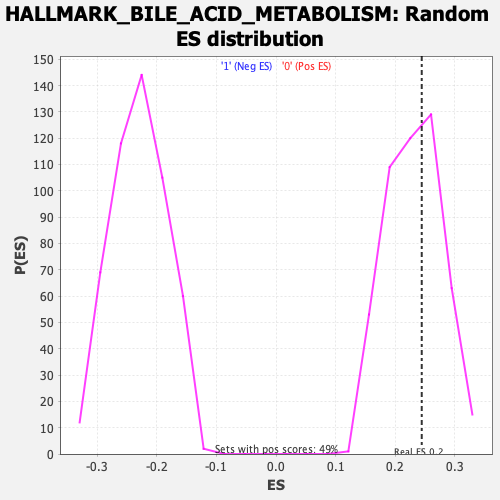
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.24480541 |
| Normalized Enrichment Score (NES) | 1.0498588 |
| Nominal p-value | 0.41632652 |
| FDR q-value | 0.8221919 |
| FWER p-Value | 0.968 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rxra | 125 | 0.828 | 0.0325 | Yes |
| 2 | Cyp46a1 | 357 | 0.666 | 0.0539 | Yes |
| 3 | Rxrg | 645 | 0.577 | 0.0694 | Yes |
| 4 | Slc22a18 | 675 | 0.573 | 0.0940 | Yes |
| 5 | Abca2 | 740 | 0.559 | 0.1167 | Yes |
| 6 | Efhc1 | 783 | 0.549 | 0.1397 | Yes |
| 7 | Dhcr24 | 792 | 0.548 | 0.1639 | Yes |
| 8 | Hsd3b7 | 839 | 0.540 | 0.1864 | Yes |
| 9 | Pex11a | 1710 | 0.433 | 0.1742 | Yes |
| 10 | Tfcp2l1 | 2597 | 0.364 | 0.1582 | Yes |
| 11 | Nr3c2 | 2682 | 0.359 | 0.1712 | Yes |
| 12 | Pex11g | 2716 | 0.356 | 0.1860 | Yes |
| 13 | Gc | 2806 | 0.351 | 0.1985 | Yes |
| 14 | Nr1h4 | 3814 | 0.295 | 0.1751 | Yes |
| 15 | Nr1i2 | 3847 | 0.294 | 0.1871 | Yes |
| 16 | Rbp1 | 4106 | 0.280 | 0.1902 | Yes |
| 17 | Aqp9 | 4321 | 0.270 | 0.1945 | Yes |
| 18 | Idi1 | 4515 | 0.261 | 0.1992 | Yes |
| 19 | Abcd1 | 4631 | 0.256 | 0.2064 | Yes |
| 20 | Pex26 | 4860 | 0.247 | 0.2092 | Yes |
| 21 | Lck | 4971 | 0.241 | 0.2160 | Yes |
| 22 | Soat2 | 5482 | 0.224 | 0.2075 | Yes |
| 23 | Nr0b2 | 5757 | 0.212 | 0.2070 | Yes |
| 24 | Cyp39a1 | 5939 | 0.205 | 0.2096 | Yes |
| 25 | Hacl1 | 6133 | 0.197 | 0.2114 | Yes |
| 26 | Pex12 | 6429 | 0.187 | 0.2090 | Yes |
| 27 | Pex7 | 6436 | 0.186 | 0.2172 | Yes |
| 28 | Pipox | 6771 | 0.174 | 0.2128 | Yes |
| 29 | Pex16 | 6831 | 0.173 | 0.2184 | Yes |
| 30 | Slc23a2 | 6898 | 0.171 | 0.2237 | Yes |
| 31 | Pnpla8 | 7092 | 0.164 | 0.2240 | Yes |
| 32 | Pecr | 7099 | 0.164 | 0.2311 | Yes |
| 33 | Pex1 | 7488 | 0.154 | 0.2239 | Yes |
| 34 | Slco1a4 | 7539 | 0.152 | 0.2289 | Yes |
| 35 | Hsd17b11 | 7668 | 0.147 | 0.2308 | Yes |
| 36 | Fads1 | 7669 | 0.147 | 0.2374 | Yes |
| 37 | Abcd3 | 7725 | 0.145 | 0.2419 | Yes |
| 38 | Phyh | 7821 | 0.142 | 0.2448 | Yes |
| 39 | Atxn1 | 8132 | 0.133 | 0.2395 | No |
| 40 | Fads2 | 8301 | 0.128 | 0.2391 | No |
| 41 | Optn | 8599 | 0.119 | 0.2336 | No |
| 42 | Gstk1 | 8898 | 0.111 | 0.2278 | No |
| 43 | Sult2b1 | 9074 | 0.105 | 0.2261 | No |
| 44 | Bcar3 | 9474 | 0.094 | 0.2158 | No |
| 45 | Lonp2 | 9804 | 0.086 | 0.2077 | No |
| 46 | Lipe | 9896 | 0.084 | 0.2081 | No |
| 47 | Idh1 | 9966 | 0.082 | 0.2093 | No |
| 48 | Mlycd | 9968 | 0.082 | 0.2129 | No |
| 49 | Nudt12 | 10079 | 0.078 | 0.2124 | No |
| 50 | Abca9 | 10125 | 0.077 | 0.2142 | No |
| 51 | Dio1 | 10157 | 0.076 | 0.2165 | No |
| 52 | Crot | 10508 | 0.067 | 0.2068 | No |
| 53 | Fdxr | 10528 | 0.066 | 0.2091 | No |
| 54 | Bbox1 | 10645 | 0.063 | 0.2077 | No |
| 55 | Npc1 | 10814 | 0.059 | 0.2042 | No |
| 56 | Abcg4 | 10896 | 0.057 | 0.2038 | No |
| 57 | Abca8b | 11029 | 0.053 | 0.2014 | No |
| 58 | Acsl1 | 11353 | 0.045 | 0.1917 | No |
| 59 | Abca3 | 11583 | 0.039 | 0.1851 | No |
| 60 | Hsd17b4 | 11623 | 0.038 | 0.1854 | No |
| 61 | Idh2 | 11639 | 0.038 | 0.1865 | No |
| 62 | Slc23a1 | 12005 | 0.031 | 0.1746 | No |
| 63 | Dio2 | 12716 | 0.015 | 0.1495 | No |
| 64 | Abca6 | 13038 | 0.007 | 0.1381 | No |
| 65 | Cat | 13981 | -0.010 | 0.1043 | No |
| 66 | Abcd2 | 14182 | -0.014 | 0.0977 | No |
| 67 | Pex19 | 14311 | -0.016 | 0.0938 | No |
| 68 | Ar | 14869 | -0.030 | 0.0748 | No |
| 69 | Abca4 | 14977 | -0.032 | 0.0724 | No |
| 70 | Gnpat | 15646 | -0.048 | 0.0502 | No |
| 71 | Aldh9a1 | 16390 | -0.065 | 0.0261 | No |
| 72 | Slc27a5 | 16908 | -0.078 | 0.0108 | No |
| 73 | Aldh1a1 | 17477 | -0.092 | -0.0057 | No |
| 74 | Acsl5 | 17792 | -0.100 | -0.0127 | No |
| 75 | Pex13 | 18104 | -0.107 | -0.0192 | No |
| 76 | Prdx5 | 18794 | -0.125 | -0.0386 | No |
| 77 | Cyp27a1 | 18867 | -0.127 | -0.0356 | No |
| 78 | Slc27a2 | 19847 | -0.149 | -0.0645 | No |
| 79 | Nedd4 | 20275 | -0.163 | -0.0727 | No |
| 80 | Apoa1 | 20432 | -0.166 | -0.0710 | No |
| 81 | Pex6 | 20507 | -0.168 | -0.0661 | No |
| 82 | Pfkm | 21207 | -0.191 | -0.0830 | No |
| 83 | Paox | 21574 | -0.205 | -0.0871 | No |
| 84 | Gnmt | 21755 | -0.211 | -0.0842 | No |
| 85 | Abca1 | 21836 | -0.215 | -0.0775 | No |
| 86 | Gclm | 21919 | -0.219 | -0.0707 | No |
| 87 | Pxmp2 | 22514 | -0.243 | -0.0814 | No |
| 88 | Cyp7b1 | 22863 | -0.258 | -0.0825 | No |
| 89 | Aldh8a1 | 22944 | -0.261 | -0.0737 | No |
| 90 | Scp2 | 23453 | -0.284 | -0.0795 | No |
| 91 | Klf1 | 23580 | -0.289 | -0.0712 | No |
| 92 | Slc29a1 | 23762 | -0.299 | -0.0644 | No |
| 93 | Retsat | 23793 | -0.301 | -0.0520 | No |
| 94 | Sod1 | 24041 | -0.317 | -0.0468 | No |
| 95 | Abca5 | 24119 | -0.318 | -0.0353 | No |
| 96 | Amacr | 24358 | -0.330 | -0.0292 | No |
| 97 | Isoc1 | 24584 | -0.342 | -0.0221 | No |
| 98 | Slc35b2 | 24754 | -0.351 | -0.0125 | No |
| 99 | Akr1d1 | 24791 | -0.354 | 0.0020 | No |
| 100 | Bmp6 | 24808 | -0.355 | 0.0173 | No |
| 101 | Ephx2 | 25382 | -0.397 | 0.0142 | No |
| 102 | Sult1b1 | 25753 | -0.430 | 0.0200 | No |
| 103 | Abcg8 | 26029 | -0.455 | 0.0304 | No |
| 104 | Hao1 | 26964 | -0.598 | 0.0232 | No |