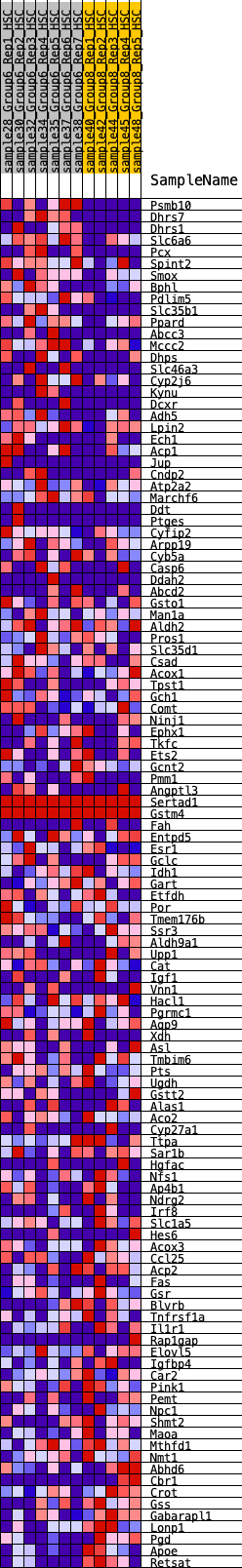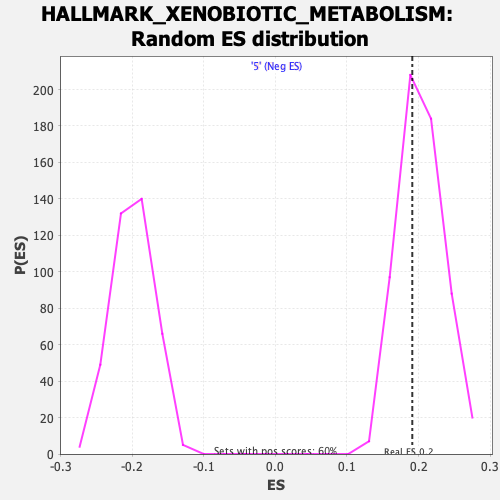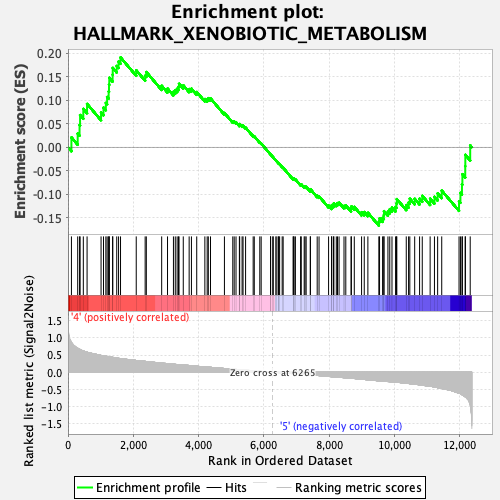
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | 0.19130935 |
| Normalized Enrichment Score (NES) | 0.93801755 |
| Nominal p-value | 0.64403975 |
| FDR q-value | 0.8462539 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Psmb10 | 105 | 0.865 | 0.0209 | Yes |
| 2 | Dhrs7 | 297 | 0.696 | 0.0291 | Yes |
| 3 | Dhrs1 | 354 | 0.669 | 0.0473 | Yes |
| 4 | Slc6a6 | 370 | 0.661 | 0.0686 | Yes |
| 5 | Pcx | 469 | 0.618 | 0.0816 | Yes |
| 6 | Spint2 | 584 | 0.584 | 0.0923 | Yes |
| 7 | Smox | 1012 | 0.489 | 0.0741 | Yes |
| 8 | Bphl | 1087 | 0.479 | 0.0843 | Yes |
| 9 | Pdlim5 | 1159 | 0.468 | 0.0945 | Yes |
| 10 | Slc35b1 | 1198 | 0.461 | 0.1071 | Yes |
| 11 | Ppard | 1245 | 0.451 | 0.1187 | Yes |
| 12 | Abcc3 | 1256 | 0.449 | 0.1332 | Yes |
| 13 | Mccc2 | 1264 | 0.448 | 0.1479 | Yes |
| 14 | Dhps | 1366 | 0.434 | 0.1545 | Yes |
| 15 | Slc46a3 | 1370 | 0.433 | 0.1690 | Yes |
| 16 | Cyp2j6 | 1492 | 0.413 | 0.1732 | Yes |
| 17 | Kynu | 1548 | 0.406 | 0.1825 | Yes |
| 18 | Dcxr | 1608 | 0.399 | 0.1913 | Yes |
| 19 | Adh5 | 2089 | 0.343 | 0.1638 | No |
| 20 | Lpin2 | 2362 | 0.317 | 0.1524 | No |
| 21 | Ech1 | 2398 | 0.311 | 0.1602 | No |
| 22 | Acp1 | 2868 | 0.265 | 0.1309 | No |
| 23 | Jup | 3047 | 0.252 | 0.1250 | No |
| 24 | Cndp2 | 3228 | 0.235 | 0.1183 | No |
| 25 | Atp2a2 | 3284 | 0.230 | 0.1216 | No |
| 26 | Marchf6 | 3341 | 0.225 | 0.1247 | No |
| 27 | Ddt | 3384 | 0.221 | 0.1288 | No |
| 28 | Ptges | 3399 | 0.221 | 0.1352 | No |
| 29 | Cyfip2 | 3529 | 0.211 | 0.1318 | No |
| 30 | Arpp19 | 3709 | 0.197 | 0.1239 | No |
| 31 | Cyb5a | 3779 | 0.190 | 0.1248 | No |
| 32 | Casp6 | 3941 | 0.177 | 0.1176 | No |
| 33 | Ddah2 | 4190 | 0.155 | 0.1027 | No |
| 34 | Abcd2 | 4261 | 0.153 | 0.1022 | No |
| 35 | Gsto1 | 4296 | 0.149 | 0.1045 | No |
| 36 | Man1a | 4363 | 0.144 | 0.1040 | No |
| 37 | Aldh2 | 4789 | 0.110 | 0.0730 | No |
| 38 | Pros1 | 5045 | 0.090 | 0.0553 | No |
| 39 | Slc35d1 | 5088 | 0.088 | 0.0548 | No |
| 40 | Csad | 5144 | 0.084 | 0.0532 | No |
| 41 | Acox1 | 5247 | 0.078 | 0.0475 | No |
| 42 | Tpst1 | 5256 | 0.076 | 0.0495 | No |
| 43 | Gch1 | 5330 | 0.070 | 0.0459 | No |
| 44 | Comt | 5359 | 0.068 | 0.0459 | No |
| 45 | Ninj1 | 5437 | 0.062 | 0.0418 | No |
| 46 | Ephx1 | 5669 | 0.044 | 0.0244 | No |
| 47 | Tkfc | 5703 | 0.041 | 0.0231 | No |
| 48 | Ets2 | 5872 | 0.029 | 0.0104 | No |
| 49 | Gcnt2 | 5914 | 0.026 | 0.0079 | No |
| 50 | Pmm1 | 6201 | 0.003 | -0.0153 | No |
| 51 | Angptl3 | 6203 | 0.003 | -0.0153 | No |
| 52 | Sertad1 | 6269 | 0.000 | -0.0206 | No |
| 53 | Gstm4 | 6284 | 0.000 | -0.0217 | No |
| 54 | Fah | 6363 | -0.001 | -0.0280 | No |
| 55 | Entpd5 | 6396 | -0.003 | -0.0305 | No |
| 56 | Esr1 | 6447 | -0.008 | -0.0344 | No |
| 57 | Gclc | 6453 | -0.008 | -0.0345 | No |
| 58 | Idh1 | 6479 | -0.010 | -0.0362 | No |
| 59 | Gart | 6557 | -0.014 | -0.0420 | No |
| 60 | Etfdh | 6587 | -0.017 | -0.0438 | No |
| 61 | Por | 6894 | -0.039 | -0.0675 | No |
| 62 | Tmem176b | 6900 | -0.039 | -0.0665 | No |
| 63 | Ssr3 | 6923 | -0.041 | -0.0669 | No |
| 64 | Aldh9a1 | 6962 | -0.045 | -0.0685 | No |
| 65 | Upp1 | 7121 | -0.057 | -0.0795 | No |
| 66 | Cat | 7141 | -0.058 | -0.0790 | No |
| 67 | Igf1 | 7231 | -0.064 | -0.0841 | No |
| 68 | Vnn1 | 7257 | -0.066 | -0.0839 | No |
| 69 | Hacl1 | 7291 | -0.068 | -0.0843 | No |
| 70 | Pgrmc1 | 7419 | -0.077 | -0.0920 | No |
| 71 | Aqp9 | 7421 | -0.077 | -0.0895 | No |
| 72 | Xdh | 7629 | -0.091 | -0.1033 | No |
| 73 | Asl | 7686 | -0.095 | -0.1046 | No |
| 74 | Tmbim6 | 7978 | -0.119 | -0.1243 | No |
| 75 | Pts | 8069 | -0.127 | -0.1274 | No |
| 76 | Ugdh | 8075 | -0.128 | -0.1234 | No |
| 77 | Gstt2 | 8130 | -0.132 | -0.1233 | No |
| 78 | Alas1 | 8139 | -0.132 | -0.1195 | No |
| 79 | Aco2 | 8214 | -0.137 | -0.1208 | No |
| 80 | Cyp27a1 | 8251 | -0.140 | -0.1190 | No |
| 81 | Ttpa | 8299 | -0.143 | -0.1180 | No |
| 82 | Sar1b | 8450 | -0.155 | -0.1249 | No |
| 83 | Hgfac | 8505 | -0.159 | -0.1239 | No |
| 84 | Nfs1 | 8666 | -0.171 | -0.1311 | No |
| 85 | Ap4b1 | 8676 | -0.172 | -0.1260 | No |
| 86 | Ndrg2 | 8765 | -0.179 | -0.1271 | No |
| 87 | Irf8 | 8988 | -0.199 | -0.1384 | No |
| 88 | Slc1a5 | 9070 | -0.206 | -0.1380 | No |
| 89 | Hes6 | 9181 | -0.217 | -0.1396 | No |
| 90 | Acox3 | 9517 | -0.248 | -0.1585 | No |
| 91 | Ccl25 | 9538 | -0.250 | -0.1516 | No |
| 92 | Acp2 | 9631 | -0.254 | -0.1504 | No |
| 93 | Fas | 9669 | -0.258 | -0.1447 | No |
| 94 | Gsr | 9674 | -0.258 | -0.1362 | No |
| 95 | Blvrb | 9795 | -0.268 | -0.1368 | No |
| 96 | Tnfrsf1a | 9851 | -0.272 | -0.1321 | No |
| 97 | Il1r1 | 9919 | -0.278 | -0.1281 | No |
| 98 | Rap1gap | 10029 | -0.287 | -0.1272 | No |
| 99 | Elovl5 | 10057 | -0.288 | -0.1196 | No |
| 100 | Igfbp4 | 10067 | -0.288 | -0.1105 | No |
| 101 | Car2 | 10356 | -0.318 | -0.1232 | No |
| 102 | Pink1 | 10427 | -0.324 | -0.1179 | No |
| 103 | Pemt | 10465 | -0.329 | -0.1097 | No |
| 104 | Npc1 | 10614 | -0.348 | -0.1099 | No |
| 105 | Shmt2 | 10764 | -0.363 | -0.1097 | No |
| 106 | Maoa | 10846 | -0.374 | -0.1035 | No |
| 107 | Mthfd1 | 11087 | -0.403 | -0.1094 | No |
| 108 | Nmt1 | 11219 | -0.425 | -0.1056 | No |
| 109 | Abhd6 | 11317 | -0.446 | -0.0983 | No |
| 110 | Cbr1 | 11442 | -0.471 | -0.0924 | No |
| 111 | Crot | 11972 | -0.607 | -0.1149 | No |
| 112 | Gss | 12015 | -0.619 | -0.0972 | No |
| 113 | Gabarapl1 | 12062 | -0.648 | -0.0789 | No |
| 114 | Lonp1 | 12075 | -0.658 | -0.0574 | No |
| 115 | Pgd | 12162 | -0.710 | -0.0402 | No |
| 116 | Apoe | 12165 | -0.712 | -0.0161 | No |
| 117 | Retsat | 12314 | -0.945 | 0.0040 | No |