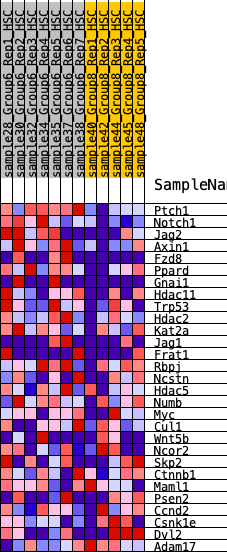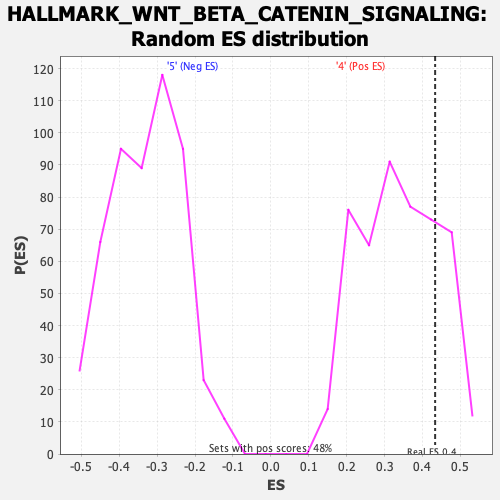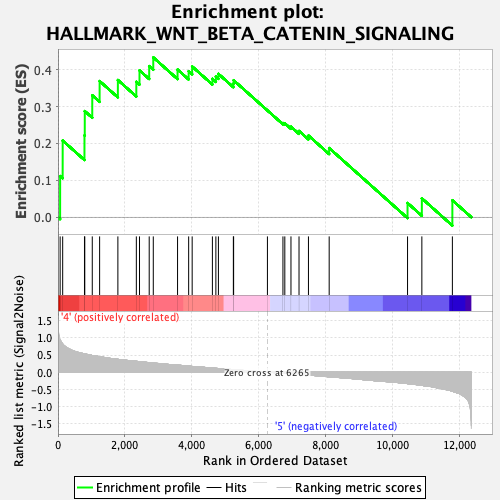
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.43343496 |
| Normalized Enrichment Score (NES) | 1.2778816 |
| Nominal p-value | 0.20754717 |
| FDR q-value | 0.68656105 |
| FWER p-Value | 0.796 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ptch1 | 64 | 0.935 | 0.1116 | Yes |
| 2 | Notch1 | 140 | 0.820 | 0.2079 | Yes |
| 3 | Jag2 | 792 | 0.534 | 0.2219 | Yes |
| 4 | Axin1 | 801 | 0.532 | 0.2878 | Yes |
| 5 | Fzd8 | 1025 | 0.488 | 0.3307 | Yes |
| 6 | Ppard | 1245 | 0.451 | 0.3692 | Yes |
| 7 | Gnai1 | 1791 | 0.376 | 0.3721 | Yes |
| 8 | Hdac11 | 2343 | 0.319 | 0.3673 | Yes |
| 9 | Trp53 | 2438 | 0.309 | 0.3983 | Yes |
| 10 | Hdac2 | 2728 | 0.279 | 0.4098 | Yes |
| 11 | Kat2a | 2850 | 0.268 | 0.4334 | Yes |
| 12 | Jag1 | 3576 | 0.207 | 0.4005 | No |
| 13 | Frat1 | 3908 | 0.179 | 0.3961 | No |
| 14 | Rbpj | 4013 | 0.170 | 0.4088 | No |
| 15 | Ncstn | 4618 | 0.123 | 0.3753 | No |
| 16 | Hdac5 | 4722 | 0.115 | 0.3813 | No |
| 17 | Numb | 4798 | 0.109 | 0.3889 | No |
| 18 | Myc | 5245 | 0.078 | 0.3624 | No |
| 19 | Cul1 | 5253 | 0.077 | 0.3715 | No |
| 20 | Wnt5b | 6264 | 0.000 | 0.2896 | No |
| 21 | Ncor2 | 6728 | -0.027 | 0.2554 | No |
| 22 | Skp2 | 6784 | -0.030 | 0.2547 | No |
| 23 | Ctnnb1 | 6968 | -0.045 | 0.2455 | No |
| 24 | Maml1 | 7209 | -0.063 | 0.2339 | No |
| 25 | Psen2 | 7491 | -0.083 | 0.2215 | No |
| 26 | Ccnd2 | 8111 | -0.130 | 0.1876 | No |
| 27 | Csnk1e | 10455 | -0.328 | 0.0386 | No |
| 28 | Dvl2 | 10883 | -0.378 | 0.0512 | No |
| 29 | Adam17 | 11796 | -0.550 | 0.0460 | No |