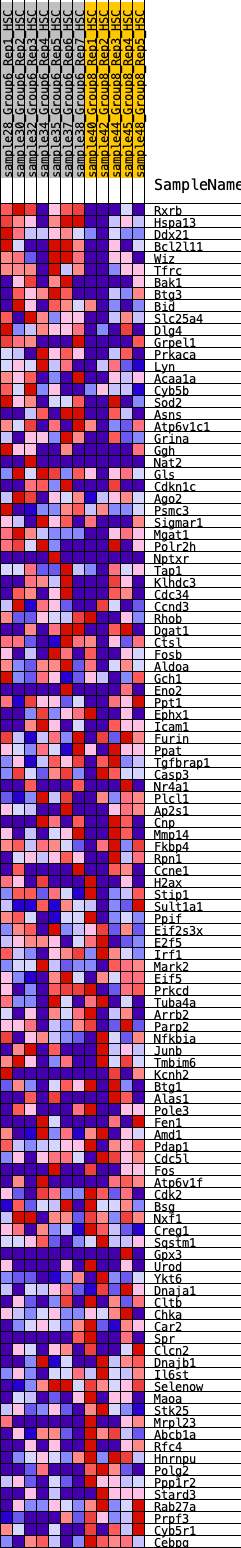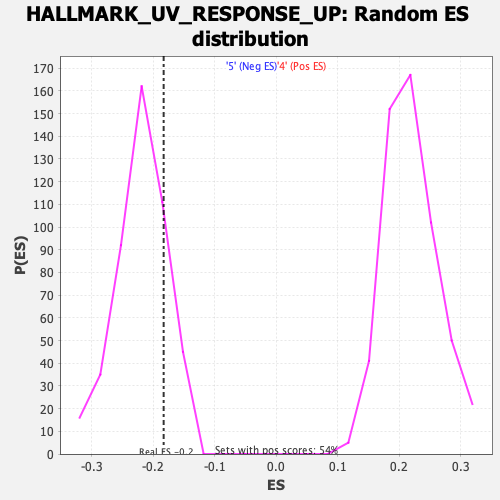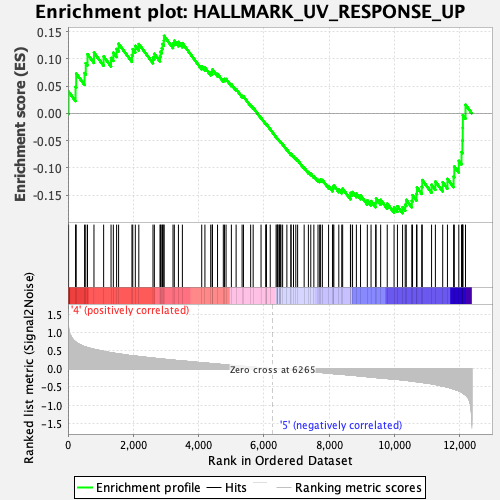
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.18327641 |
| Normalized Enrichment Score (NES) | -0.8323745 |
| Nominal p-value | 0.8199566 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rxrb | 18 | 1.156 | 0.0398 | No |
| 2 | Hspa13 | 231 | 0.737 | 0.0487 | No |
| 3 | Ddx21 | 251 | 0.724 | 0.0730 | No |
| 4 | Bcl2l11 | 507 | 0.606 | 0.0738 | No |
| 5 | Wiz | 544 | 0.596 | 0.0921 | No |
| 6 | Tfrc | 597 | 0.579 | 0.1085 | No |
| 7 | Bak1 | 795 | 0.534 | 0.1115 | No |
| 8 | Btg3 | 1091 | 0.478 | 0.1044 | No |
| 9 | Bid | 1318 | 0.441 | 0.1017 | No |
| 10 | Slc25a4 | 1388 | 0.430 | 0.1114 | No |
| 11 | Dlg4 | 1488 | 0.414 | 0.1181 | No |
| 12 | Grpel1 | 1549 | 0.405 | 0.1277 | No |
| 13 | Prkaca | 1958 | 0.358 | 0.1072 | No |
| 14 | Lyn | 1983 | 0.354 | 0.1178 | No |
| 15 | Acaa1a | 2061 | 0.347 | 0.1239 | No |
| 16 | Cyb5b | 2168 | 0.336 | 0.1272 | No |
| 17 | Sod2 | 2602 | 0.293 | 0.1024 | No |
| 18 | Asns | 2644 | 0.288 | 0.1093 | No |
| 19 | Atp6v1c1 | 2817 | 0.271 | 0.1049 | No |
| 20 | Grina | 2834 | 0.269 | 0.1132 | No |
| 21 | Ggh | 2873 | 0.264 | 0.1195 | No |
| 22 | Nat2 | 2894 | 0.263 | 0.1272 | No |
| 23 | Gls | 2934 | 0.260 | 0.1333 | No |
| 24 | Cdkn1c | 2940 | 0.259 | 0.1422 | No |
| 25 | Ago2 | 3215 | 0.236 | 0.1282 | No |
| 26 | Psmc3 | 3255 | 0.233 | 0.1333 | No |
| 27 | Sigmar1 | 3381 | 0.221 | 0.1310 | No |
| 28 | Mgat1 | 3502 | 0.213 | 0.1288 | No |
| 29 | Polr2h | 4094 | 0.163 | 0.0864 | No |
| 30 | Nptxr | 4192 | 0.155 | 0.0840 | No |
| 31 | Tap1 | 4369 | 0.143 | 0.0748 | No |
| 32 | Klhdc3 | 4415 | 0.139 | 0.0761 | No |
| 33 | Cdc34 | 4422 | 0.139 | 0.0805 | No |
| 34 | Ccnd3 | 4578 | 0.126 | 0.0724 | No |
| 35 | Rhob | 4755 | 0.112 | 0.0620 | No |
| 36 | Dgat1 | 4790 | 0.110 | 0.0632 | No |
| 37 | Ctsl | 4839 | 0.106 | 0.0630 | No |
| 38 | Fosb | 5002 | 0.094 | 0.0532 | No |
| 39 | Aldoa | 5147 | 0.084 | 0.0444 | No |
| 40 | Gch1 | 5330 | 0.070 | 0.0321 | No |
| 41 | Eno2 | 5372 | 0.067 | 0.0311 | No |
| 42 | Ppt1 | 5592 | 0.050 | 0.0150 | No |
| 43 | Ephx1 | 5669 | 0.044 | 0.0104 | No |
| 44 | Icam1 | 5910 | 0.026 | -0.0083 | No |
| 45 | Furin | 6055 | 0.015 | -0.0195 | No |
| 46 | Ppat | 6069 | 0.014 | -0.0201 | No |
| 47 | Tgfbrap1 | 6074 | 0.014 | -0.0199 | No |
| 48 | Casp3 | 6192 | 0.004 | -0.0293 | No |
| 49 | Nr4a1 | 6371 | -0.002 | -0.0438 | No |
| 50 | Plcl1 | 6401 | -0.004 | -0.0460 | No |
| 51 | Ap2s1 | 6422 | -0.006 | -0.0474 | No |
| 52 | Cnp | 6423 | -0.006 | -0.0472 | No |
| 53 | Mmp14 | 6466 | -0.009 | -0.0503 | No |
| 54 | Fkbp4 | 6478 | -0.009 | -0.0509 | No |
| 55 | Rpn1 | 6513 | -0.011 | -0.0533 | No |
| 56 | Ccne1 | 6515 | -0.012 | -0.0529 | No |
| 57 | H2ax | 6571 | -0.015 | -0.0569 | No |
| 58 | Stip1 | 6701 | -0.025 | -0.0665 | No |
| 59 | Sult1a1 | 6826 | -0.034 | -0.0754 | No |
| 60 | Ppif | 6827 | -0.034 | -0.0742 | No |
| 61 | Eif2s3x | 6904 | -0.040 | -0.0790 | No |
| 62 | E2f5 | 6974 | -0.046 | -0.0830 | No |
| 63 | Irf1 | 7030 | -0.049 | -0.0857 | No |
| 64 | Mark2 | 7230 | -0.064 | -0.0997 | No |
| 65 | Eif5 | 7360 | -0.073 | -0.1076 | No |
| 66 | Prkcd | 7433 | -0.078 | -0.1107 | No |
| 67 | Tuba4a | 7527 | -0.085 | -0.1153 | No |
| 68 | Arrb2 | 7654 | -0.093 | -0.1222 | No |
| 69 | Parp2 | 7705 | -0.096 | -0.1229 | No |
| 70 | Nfkbia | 7734 | -0.098 | -0.1217 | No |
| 71 | Junb | 7788 | -0.102 | -0.1223 | No |
| 72 | Tmbim6 | 7978 | -0.119 | -0.1335 | No |
| 73 | Kcnh2 | 8097 | -0.129 | -0.1386 | No |
| 74 | Btg1 | 8115 | -0.131 | -0.1353 | No |
| 75 | Alas1 | 8139 | -0.132 | -0.1324 | No |
| 76 | Pole3 | 8291 | -0.142 | -0.1397 | No |
| 77 | Fen1 | 8381 | -0.149 | -0.1416 | No |
| 78 | Amd1 | 8409 | -0.152 | -0.1384 | No |
| 79 | Pdap1 | 8647 | -0.171 | -0.1517 | No |
| 80 | Cdc5l | 8654 | -0.171 | -0.1461 | No |
| 81 | Fos | 8712 | -0.174 | -0.1445 | No |
| 82 | Atp6v1f | 8829 | -0.185 | -0.1473 | No |
| 83 | Cdk2 | 8956 | -0.196 | -0.1506 | No |
| 84 | Bsg | 9164 | -0.215 | -0.1598 | No |
| 85 | Nxf1 | 9278 | -0.225 | -0.1610 | No |
| 86 | Creg1 | 9420 | -0.238 | -0.1641 | No |
| 87 | Sqstm1 | 9431 | -0.239 | -0.1564 | No |
| 88 | Gpx3 | 9572 | -0.252 | -0.1588 | No |
| 89 | Urod | 9774 | -0.267 | -0.1657 | No |
| 90 | Ykt6 | 9983 | -0.283 | -0.1726 | Yes |
| 91 | Dnaja1 | 10086 | -0.290 | -0.1705 | Yes |
| 92 | Cltb | 10243 | -0.306 | -0.1724 | Yes |
| 93 | Chka | 10324 | -0.314 | -0.1677 | Yes |
| 94 | Car2 | 10356 | -0.318 | -0.1589 | Yes |
| 95 | Spr | 10526 | -0.337 | -0.1607 | Yes |
| 96 | Clcn2 | 10548 | -0.339 | -0.1503 | Yes |
| 97 | Dnajb1 | 10669 | -0.352 | -0.1475 | Yes |
| 98 | Il6st | 10684 | -0.353 | -0.1361 | Yes |
| 99 | Selenow | 10830 | -0.371 | -0.1346 | Yes |
| 100 | Maoa | 10846 | -0.374 | -0.1225 | Yes |
| 101 | Stk25 | 11131 | -0.410 | -0.1311 | Yes |
| 102 | Mrpl23 | 11248 | -0.430 | -0.1252 | Yes |
| 103 | Abcb1a | 11472 | -0.474 | -0.1265 | Yes |
| 104 | Rfc4 | 11617 | -0.503 | -0.1203 | Yes |
| 105 | Hnrnpu | 11806 | -0.553 | -0.1159 | Yes |
| 106 | Polg2 | 11827 | -0.559 | -0.0976 | Yes |
| 107 | Ppp1r2 | 11965 | -0.605 | -0.0872 | Yes |
| 108 | Stard3 | 12048 | -0.641 | -0.0710 | Yes |
| 109 | Rab27a | 12076 | -0.658 | -0.0498 | Yes |
| 110 | Prpf3 | 12085 | -0.661 | -0.0268 | Yes |
| 111 | Cyb5r1 | 12089 | -0.665 | -0.0033 | Yes |
| 112 | Cebpg | 12170 | -0.718 | 0.0158 | Yes |