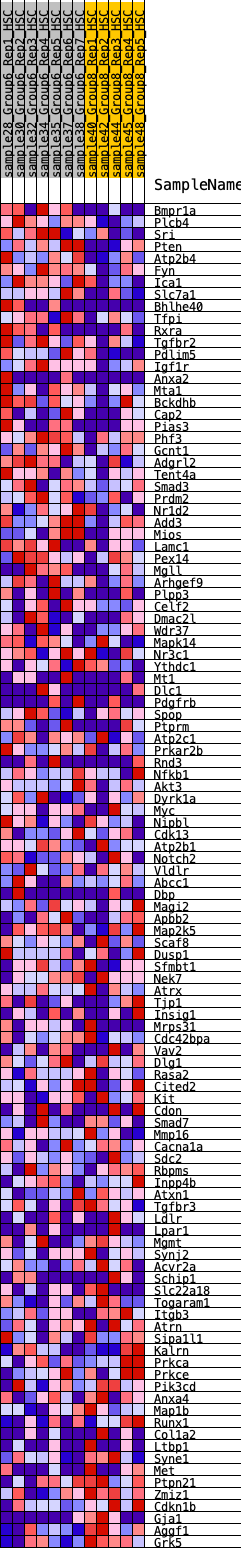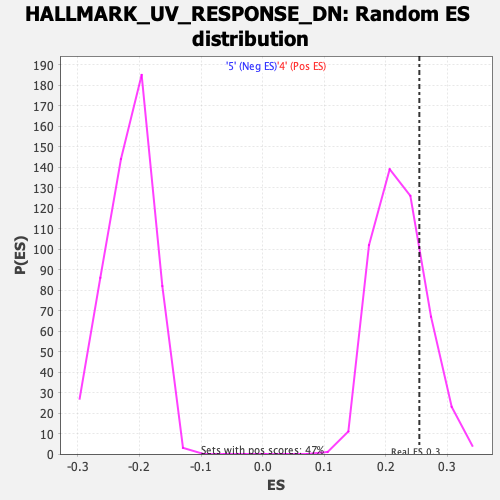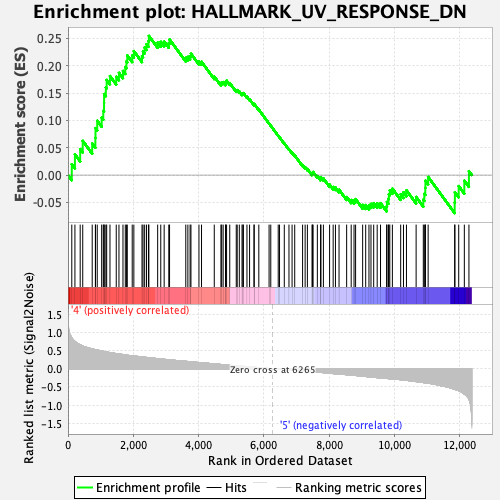
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.25459167 |
| Normalized Enrichment Score (NES) | 1.145393 |
| Nominal p-value | 0.20718816 |
| FDR q-value | 0.5886711 |
| FWER p-Value | 0.973 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bmpr1a | 116 | 0.854 | 0.0199 | Yes |
| 2 | Plcb4 | 212 | 0.753 | 0.0380 | Yes |
| 3 | Sri | 373 | 0.659 | 0.0475 | Yes |
| 4 | Pten | 451 | 0.623 | 0.0626 | Yes |
| 5 | Atp2b4 | 741 | 0.546 | 0.0578 | Yes |
| 6 | Fyn | 838 | 0.527 | 0.0680 | Yes |
| 7 | Ica1 | 842 | 0.527 | 0.0859 | Yes |
| 8 | Slc7a1 | 895 | 0.516 | 0.0993 | Yes |
| 9 | Bhlhe40 | 1031 | 0.488 | 0.1051 | Yes |
| 10 | Tfpi | 1080 | 0.479 | 0.1176 | Yes |
| 11 | Rxra | 1104 | 0.476 | 0.1320 | Yes |
| 12 | Tgfbr2 | 1106 | 0.476 | 0.1483 | Yes |
| 13 | Pdlim5 | 1159 | 0.468 | 0.1601 | Yes |
| 14 | Igf1r | 1184 | 0.463 | 0.1740 | Yes |
| 15 | Anxa2 | 1285 | 0.445 | 0.1812 | Yes |
| 16 | Mta1 | 1477 | 0.416 | 0.1798 | Yes |
| 17 | Bckdhb | 1560 | 0.403 | 0.1870 | Yes |
| 18 | Cap2 | 1683 | 0.391 | 0.1904 | Yes |
| 19 | Pias3 | 1754 | 0.382 | 0.1978 | Yes |
| 20 | Phf3 | 1789 | 0.377 | 0.2080 | Yes |
| 21 | Gcnt1 | 1815 | 0.373 | 0.2188 | Yes |
| 22 | Adgrl2 | 1966 | 0.356 | 0.2187 | Yes |
| 23 | Tent4a | 2016 | 0.351 | 0.2268 | Yes |
| 24 | Smad3 | 2266 | 0.326 | 0.2176 | Yes |
| 25 | Prdm2 | 2297 | 0.323 | 0.2263 | Yes |
| 26 | Nr1d2 | 2347 | 0.319 | 0.2332 | Yes |
| 27 | Add3 | 2404 | 0.311 | 0.2393 | Yes |
| 28 | Mios | 2464 | 0.306 | 0.2450 | Yes |
| 29 | Lamc1 | 2476 | 0.305 | 0.2546 | Yes |
| 30 | Pex14 | 2745 | 0.278 | 0.2423 | No |
| 31 | Mgll | 2840 | 0.268 | 0.2438 | No |
| 32 | Arhgef9 | 2944 | 0.259 | 0.2443 | No |
| 33 | Plpp3 | 3086 | 0.250 | 0.2414 | No |
| 34 | Celf2 | 3109 | 0.248 | 0.2481 | No |
| 35 | Dmac2l | 3606 | 0.204 | 0.2146 | No |
| 36 | Wdr37 | 3670 | 0.201 | 0.2164 | No |
| 37 | Mapk14 | 3738 | 0.194 | 0.2176 | No |
| 38 | Nr3c1 | 3764 | 0.191 | 0.2221 | No |
| 39 | Ythdc1 | 4009 | 0.170 | 0.2080 | No |
| 40 | Mt1 | 4088 | 0.164 | 0.2072 | No |
| 41 | Dlc1 | 4478 | 0.134 | 0.1801 | No |
| 42 | Pdgfrb | 4682 | 0.118 | 0.1676 | No |
| 43 | Spop | 4705 | 0.117 | 0.1698 | No |
| 44 | Ptprm | 4750 | 0.113 | 0.1701 | No |
| 45 | Atp2c1 | 4822 | 0.107 | 0.1680 | No |
| 46 | Prkar2b | 4832 | 0.107 | 0.1709 | No |
| 47 | Rnd3 | 4855 | 0.105 | 0.1727 | No |
| 48 | Nfkb1 | 4952 | 0.098 | 0.1682 | No |
| 49 | Akt3 | 5151 | 0.084 | 0.1549 | No |
| 50 | Dyrk1a | 5180 | 0.081 | 0.1554 | No |
| 51 | Myc | 5245 | 0.078 | 0.1529 | No |
| 52 | Nipbl | 5328 | 0.071 | 0.1486 | No |
| 53 | Cdk13 | 5355 | 0.068 | 0.1488 | No |
| 54 | Atp2b1 | 5370 | 0.067 | 0.1500 | No |
| 55 | Notch2 | 5479 | 0.058 | 0.1432 | No |
| 56 | Vldlr | 5559 | 0.053 | 0.1385 | No |
| 57 | Abcc1 | 5696 | 0.042 | 0.1289 | No |
| 58 | Dbp | 5699 | 0.042 | 0.1301 | No |
| 59 | Magi2 | 5842 | 0.031 | 0.1196 | No |
| 60 | Apbb2 | 6156 | 0.007 | 0.0943 | No |
| 61 | Map2k5 | 6208 | 0.003 | 0.0902 | No |
| 62 | Scaf8 | 6436 | -0.007 | 0.0719 | No |
| 63 | Dusp1 | 6476 | -0.009 | 0.0691 | No |
| 64 | Sfmbt1 | 6622 | -0.019 | 0.0579 | No |
| 65 | Nek7 | 6765 | -0.029 | 0.0473 | No |
| 66 | Atrx | 6860 | -0.037 | 0.0409 | No |
| 67 | Tjp1 | 6941 | -0.043 | 0.0358 | No |
| 68 | Insig1 | 7182 | -0.061 | 0.0183 | No |
| 69 | Mrps31 | 7260 | -0.066 | 0.0143 | No |
| 70 | Cdc42bpa | 7329 | -0.071 | 0.0112 | No |
| 71 | Vav2 | 7471 | -0.081 | 0.0025 | No |
| 72 | Dlg1 | 7487 | -0.082 | 0.0041 | No |
| 73 | Rasa2 | 7503 | -0.084 | 0.0057 | No |
| 74 | Cited2 | 7634 | -0.091 | -0.0017 | No |
| 75 | Kit | 7732 | -0.098 | -0.0063 | No |
| 76 | Cdon | 7744 | -0.099 | -0.0038 | No |
| 77 | Smad7 | 7819 | -0.105 | -0.0062 | No |
| 78 | Mmp16 | 8009 | -0.121 | -0.0175 | No |
| 79 | Cacna1a | 8118 | -0.131 | -0.0218 | No |
| 80 | Sdc2 | 8188 | -0.136 | -0.0228 | No |
| 81 | Rbpms | 8298 | -0.143 | -0.0267 | No |
| 82 | Inpp4b | 8531 | -0.160 | -0.0402 | No |
| 83 | Atxn1 | 8670 | -0.172 | -0.0455 | No |
| 84 | Tgfbr3 | 8757 | -0.179 | -0.0464 | No |
| 85 | Ldlr | 8805 | -0.183 | -0.0440 | No |
| 86 | Lpar1 | 9021 | -0.203 | -0.0546 | No |
| 87 | Mgmt | 9114 | -0.211 | -0.0548 | No |
| 88 | Synj2 | 9214 | -0.219 | -0.0554 | No |
| 89 | Acvr2a | 9276 | -0.225 | -0.0526 | No |
| 90 | Schip1 | 9359 | -0.232 | -0.0514 | No |
| 91 | Slc22a18 | 9466 | -0.243 | -0.0517 | No |
| 92 | Togaram1 | 9568 | -0.252 | -0.0513 | No |
| 93 | Itgb3 | 9755 | -0.265 | -0.0574 | No |
| 94 | Atrn | 9765 | -0.266 | -0.0490 | No |
| 95 | Sipa1l1 | 9811 | -0.269 | -0.0435 | No |
| 96 | Kalrn | 9824 | -0.269 | -0.0352 | No |
| 97 | Prkca | 9852 | -0.272 | -0.0280 | No |
| 98 | Prkce | 9931 | -0.279 | -0.0248 | No |
| 99 | Pik3cd | 10185 | -0.300 | -0.0352 | No |
| 100 | Anxa4 | 10271 | -0.309 | -0.0315 | No |
| 101 | Map1b | 10362 | -0.318 | -0.0279 | No |
| 102 | Runx1 | 10658 | -0.351 | -0.0400 | No |
| 103 | Col1a2 | 10878 | -0.378 | -0.0449 | No |
| 104 | Ltbp1 | 10909 | -0.382 | -0.0342 | No |
| 105 | Syne1 | 10939 | -0.386 | -0.0233 | No |
| 106 | Met | 10943 | -0.387 | -0.0103 | No |
| 107 | Ptpn21 | 11026 | -0.396 | -0.0034 | No |
| 108 | Zmiz1 | 11839 | -0.563 | -0.0503 | No |
| 109 | Cdkn1b | 11845 | -0.564 | -0.0314 | No |
| 110 | Gja1 | 11962 | -0.603 | -0.0201 | No |
| 111 | Aggf1 | 12134 | -0.694 | -0.0103 | No |
| 112 | Grk5 | 12275 | -0.841 | 0.0072 | No |