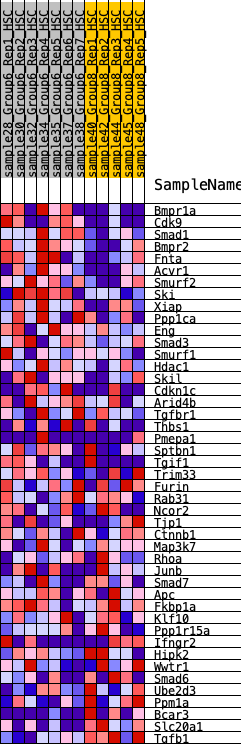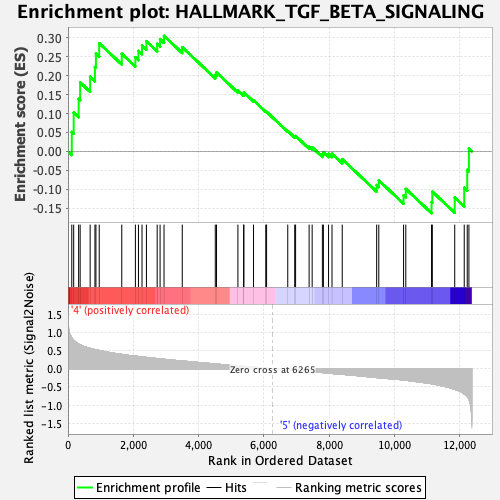
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.3046926 |
| Normalized Enrichment Score (NES) | 1.1416178 |
| Nominal p-value | 0.21824686 |
| FDR q-value | 0.54417497 |
| FWER p-Value | 0.974 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bmpr1a | 116 | 0.854 | 0.0514 | Yes |
| 2 | Cdk9 | 174 | 0.782 | 0.1024 | Yes |
| 3 | Smad1 | 326 | 0.681 | 0.1386 | Yes |
| 4 | Bmpr2 | 372 | 0.659 | 0.1819 | Yes |
| 5 | Fnta | 678 | 0.560 | 0.1970 | Yes |
| 6 | Acvr1 | 825 | 0.529 | 0.2228 | Yes |
| 7 | Smurf2 | 855 | 0.523 | 0.2576 | Yes |
| 8 | Ski | 956 | 0.502 | 0.2853 | Yes |
| 9 | Xiap | 1646 | 0.396 | 0.2575 | Yes |
| 10 | Ppp1ca | 2063 | 0.346 | 0.2484 | Yes |
| 11 | Eng | 2157 | 0.338 | 0.2649 | Yes |
| 12 | Smad3 | 2266 | 0.326 | 0.2793 | Yes |
| 13 | Smurf1 | 2401 | 0.311 | 0.2906 | Yes |
| 14 | Hdac1 | 2730 | 0.279 | 0.2838 | Yes |
| 15 | Skil | 2819 | 0.271 | 0.2960 | Yes |
| 16 | Cdkn1c | 2940 | 0.259 | 0.3047 | Yes |
| 17 | Arid4b | 3498 | 0.213 | 0.2747 | No |
| 18 | Tgfbr1 | 4509 | 0.132 | 0.2021 | No |
| 19 | Thbs1 | 4547 | 0.129 | 0.2082 | No |
| 20 | Pmepa1 | 5202 | 0.080 | 0.1608 | No |
| 21 | Sptbn1 | 5378 | 0.066 | 0.1514 | No |
| 22 | Tgif1 | 5384 | 0.066 | 0.1556 | No |
| 23 | Trim33 | 5680 | 0.043 | 0.1348 | No |
| 24 | Furin | 6055 | 0.015 | 0.1055 | No |
| 25 | Rab31 | 6080 | 0.013 | 0.1044 | No |
| 26 | Ncor2 | 6728 | -0.027 | 0.0538 | No |
| 27 | Tjp1 | 6941 | -0.043 | 0.0396 | No |
| 28 | Ctnnb1 | 6968 | -0.045 | 0.0408 | No |
| 29 | Map3k7 | 7382 | -0.075 | 0.0126 | No |
| 30 | Rhoa | 7475 | -0.081 | 0.0109 | No |
| 31 | Junb | 7788 | -0.102 | -0.0071 | No |
| 32 | Smad7 | 7819 | -0.105 | -0.0021 | No |
| 33 | Apc | 7977 | -0.119 | -0.0064 | No |
| 34 | Fkbp1a | 8083 | -0.128 | -0.0058 | No |
| 35 | Klf10 | 8396 | -0.151 | -0.0204 | No |
| 36 | Ppp1r15a | 9450 | -0.241 | -0.0887 | No |
| 37 | Ifngr2 | 9515 | -0.248 | -0.0763 | No |
| 38 | Hipk2 | 10274 | -0.309 | -0.1159 | No |
| 39 | Wwtr1 | 10342 | -0.317 | -0.0988 | No |
| 40 | Smad6 | 11130 | -0.410 | -0.1335 | No |
| 41 | Ube2d3 | 11154 | -0.414 | -0.1059 | No |
| 42 | Ppm1a | 11840 | -0.564 | -0.1214 | No |
| 43 | Bcar3 | 12131 | -0.692 | -0.0956 | No |
| 44 | Slc20a1 | 12223 | -0.766 | -0.0485 | No |
| 45 | Tgfb1 | 12274 | -0.840 | 0.0072 | No |