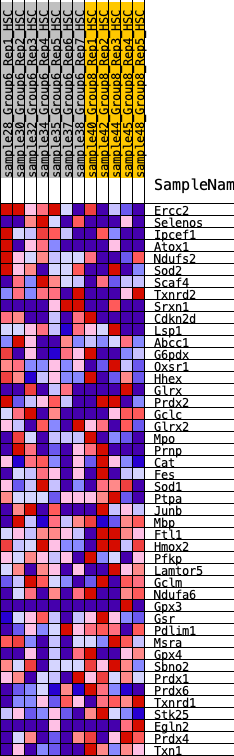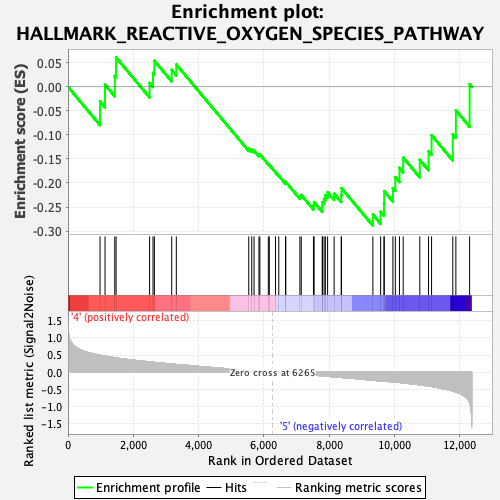
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | -0.2885895 |
| Normalized Enrichment Score (NES) | -1.089719 |
| Nominal p-value | 0.32 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ercc2 | 981 | 0.498 | -0.0300 | No |
| 2 | Selenos | 1134 | 0.471 | 0.0047 | No |
| 3 | Ipcef1 | 1432 | 0.423 | 0.0227 | No |
| 4 | Atox1 | 1472 | 0.417 | 0.0611 | No |
| 5 | Ndufs2 | 2498 | 0.303 | 0.0081 | No |
| 6 | Sod2 | 2602 | 0.293 | 0.0290 | No |
| 7 | Scaf4 | 2645 | 0.287 | 0.0542 | No |
| 8 | Txnrd2 | 3175 | 0.241 | 0.0353 | No |
| 9 | Srxn1 | 3316 | 0.227 | 0.0466 | No |
| 10 | Cdkn2d | 5534 | 0.054 | -0.1280 | No |
| 11 | Lsp1 | 5626 | 0.047 | -0.1307 | No |
| 12 | Abcc1 | 5696 | 0.042 | -0.1321 | No |
| 13 | G6pdx | 5846 | 0.031 | -0.1412 | No |
| 14 | Oxsr1 | 5874 | 0.029 | -0.1405 | No |
| 15 | Hhex | 6133 | 0.007 | -0.1607 | No |
| 16 | Glrx | 6163 | 0.006 | -0.1624 | No |
| 17 | Prdx2 | 6353 | -0.000 | -0.1777 | No |
| 18 | Gclc | 6453 | -0.008 | -0.1850 | No |
| 19 | Glrx2 | 6663 | -0.022 | -0.1998 | No |
| 20 | Mpo | 6665 | -0.022 | -0.1976 | No |
| 21 | Prnp | 7100 | -0.055 | -0.2274 | No |
| 22 | Cat | 7141 | -0.058 | -0.2248 | No |
| 23 | Fes | 7515 | -0.084 | -0.2467 | No |
| 24 | Sod1 | 7535 | -0.085 | -0.2397 | No |
| 25 | Ptpa | 7786 | -0.102 | -0.2498 | No |
| 26 | Junb | 7788 | -0.102 | -0.2397 | No |
| 27 | Mbp | 7840 | -0.107 | -0.2332 | No |
| 28 | Ftl1 | 7880 | -0.110 | -0.2254 | No |
| 29 | Hmox2 | 7948 | -0.116 | -0.2192 | No |
| 30 | Pfkp | 8148 | -0.132 | -0.2222 | No |
| 31 | Lamtor5 | 8364 | -0.148 | -0.2249 | No |
| 32 | Gclm | 8374 | -0.149 | -0.2107 | No |
| 33 | Ndufa6 | 9334 | -0.230 | -0.2657 | Yes |
| 34 | Gpx3 | 9572 | -0.252 | -0.2598 | Yes |
| 35 | Gsr | 9674 | -0.258 | -0.2422 | Yes |
| 36 | Pdlim1 | 9682 | -0.259 | -0.2170 | Yes |
| 37 | Msra | 9947 | -0.280 | -0.2104 | Yes |
| 38 | Gpx4 | 10021 | -0.287 | -0.1878 | Yes |
| 39 | Sbno2 | 10148 | -0.295 | -0.1685 | Yes |
| 40 | Prdx1 | 10262 | -0.308 | -0.1470 | Yes |
| 41 | Prdx6 | 10771 | -0.364 | -0.1519 | Yes |
| 42 | Txnrd1 | 11038 | -0.397 | -0.1339 | Yes |
| 43 | Stk25 | 11131 | -0.410 | -0.1004 | Yes |
| 44 | Egln2 | 11781 | -0.546 | -0.0987 | Yes |
| 45 | Prdx4 | 11875 | -0.576 | -0.0488 | Yes |
| 46 | Txn1 | 12296 | -0.885 | 0.0054 | Yes |