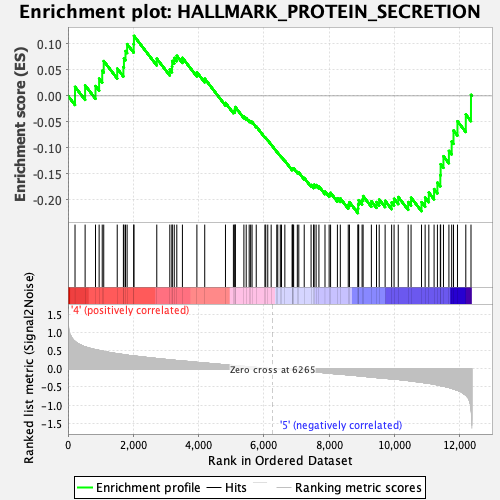
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.225373 |
| Normalized Enrichment Score (NES) | -0.9927075 |
| Nominal p-value | 0.44444445 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bnip3 | 215 | 0.750 | 0.0174 | No |
| 2 | Gbf1 | 524 | 0.600 | 0.0203 | No |
| 3 | Ica1 | 842 | 0.527 | 0.0190 | No |
| 4 | Lman1 | 953 | 0.503 | 0.0334 | No |
| 5 | Arfgef1 | 1048 | 0.487 | 0.0484 | No |
| 6 | Arf1 | 1094 | 0.477 | 0.0670 | No |
| 7 | Vps45 | 1507 | 0.411 | 0.0526 | No |
| 8 | Tsg101 | 1696 | 0.389 | 0.0554 | No |
| 9 | Sec24d | 1713 | 0.388 | 0.0721 | No |
| 10 | Gosr2 | 1760 | 0.381 | 0.0861 | No |
| 11 | Rab9 | 1810 | 0.373 | 0.0995 | No |
| 12 | Atp7a | 2011 | 0.352 | 0.0996 | No |
| 13 | Scrn1 | 2019 | 0.351 | 0.1154 | No |
| 14 | Scamp3 | 2718 | 0.280 | 0.0716 | No |
| 15 | M6pr | 3117 | 0.247 | 0.0506 | No |
| 16 | Rab2a | 3184 | 0.240 | 0.0564 | No |
| 17 | Cln5 | 3190 | 0.239 | 0.0672 | No |
| 18 | Stx7 | 3253 | 0.233 | 0.0730 | No |
| 19 | Sec22b | 3331 | 0.226 | 0.0772 | No |
| 20 | Rer1 | 3503 | 0.213 | 0.0732 | No |
| 21 | Uso1 | 3946 | 0.176 | 0.0454 | No |
| 22 | Sec31a | 4186 | 0.155 | 0.0331 | No |
| 23 | Kif1b | 4821 | 0.108 | -0.0135 | No |
| 24 | Snap23 | 5064 | 0.089 | -0.0291 | No |
| 25 | Napa | 5107 | 0.087 | -0.0285 | No |
| 26 | Vamp4 | 5115 | 0.086 | -0.0250 | No |
| 27 | Cope | 5120 | 0.086 | -0.0213 | No |
| 28 | Scamp1 | 5385 | 0.066 | -0.0398 | No |
| 29 | Napg | 5456 | 0.061 | -0.0427 | No |
| 30 | Anp32e | 5547 | 0.053 | -0.0475 | No |
| 31 | Ppt1 | 5592 | 0.050 | -0.0488 | No |
| 32 | Ergic3 | 5635 | 0.046 | -0.0501 | No |
| 33 | Rab22a | 5763 | 0.037 | -0.0587 | No |
| 34 | Ap1g1 | 6029 | 0.018 | -0.0795 | No |
| 35 | Zw10 | 6054 | 0.015 | -0.0807 | No |
| 36 | Atp1a1 | 6116 | 0.009 | -0.0852 | No |
| 37 | Dop1a | 6220 | 0.002 | -0.0935 | No |
| 38 | Dnm1l | 6392 | -0.003 | -0.1073 | No |
| 39 | Ap2s1 | 6422 | -0.006 | -0.1094 | No |
| 40 | Vamp3 | 6500 | -0.011 | -0.1152 | No |
| 41 | Ap2b1 | 6537 | -0.013 | -0.1175 | No |
| 42 | Sspn | 6639 | -0.020 | -0.1248 | No |
| 43 | Snx2 | 6863 | -0.037 | -0.1412 | No |
| 44 | Copb1 | 6870 | -0.037 | -0.1400 | No |
| 45 | Lamp2 | 6888 | -0.038 | -0.1396 | No |
| 46 | Cltc | 6908 | -0.040 | -0.1393 | No |
| 47 | Ocrl | 7021 | -0.049 | -0.1461 | No |
| 48 | Mapk1 | 7066 | -0.052 | -0.1473 | No |
| 49 | Arfip1 | 7232 | -0.064 | -0.1577 | No |
| 50 | Igf2r | 7442 | -0.078 | -0.1711 | No |
| 51 | Ctsc | 7519 | -0.084 | -0.1734 | No |
| 52 | Sod1 | 7535 | -0.085 | -0.1706 | No |
| 53 | Sgms1 | 7596 | -0.089 | -0.1713 | No |
| 54 | Tmed10 | 7682 | -0.095 | -0.1739 | No |
| 55 | Rab5a | 7868 | -0.108 | -0.1839 | No |
| 56 | Ap3b1 | 7995 | -0.120 | -0.1886 | No |
| 57 | Tmx1 | 8039 | -0.123 | -0.1863 | No |
| 58 | Clta | 8248 | -0.140 | -0.1967 | No |
| 59 | Yipf6 | 8337 | -0.146 | -0.1971 | No |
| 60 | Golga4 | 8580 | -0.164 | -0.2091 | No |
| 61 | Cav2 | 8617 | -0.168 | -0.2043 | No |
| 62 | Stam | 8877 | -0.189 | -0.2166 | Yes |
| 63 | Tpd52 | 8884 | -0.190 | -0.2082 | Yes |
| 64 | Stx16 | 8900 | -0.191 | -0.2005 | Yes |
| 65 | Cd63 | 9004 | -0.201 | -0.1995 | Yes |
| 66 | Ap2m1 | 9035 | -0.204 | -0.1925 | Yes |
| 67 | Ap3s1 | 9288 | -0.226 | -0.2025 | Yes |
| 68 | Copb2 | 9445 | -0.240 | -0.2040 | Yes |
| 69 | Tom1l1 | 9528 | -0.249 | -0.1991 | Yes |
| 70 | Adam10 | 9709 | -0.261 | -0.2016 | Yes |
| 71 | Rab14 | 9908 | -0.277 | -0.2049 | Yes |
| 72 | Ykt6 | 9983 | -0.283 | -0.1977 | Yes |
| 73 | Arcn1 | 10111 | -0.293 | -0.1944 | Yes |
| 74 | Abca1 | 10415 | -0.323 | -0.2040 | Yes |
| 75 | Arfgap3 | 10503 | -0.335 | -0.1955 | Yes |
| 76 | Bet1 | 10823 | -0.371 | -0.2043 | Yes |
| 77 | Krt18 | 10934 | -0.386 | -0.1952 | Yes |
| 78 | Galc | 11047 | -0.398 | -0.1858 | Yes |
| 79 | Atp6v1h | 11208 | -0.424 | -0.1791 | Yes |
| 80 | Arfgef2 | 11310 | -0.444 | -0.1667 | Yes |
| 81 | Dst | 11399 | -0.463 | -0.1523 | Yes |
| 82 | Gnas | 11409 | -0.466 | -0.1313 | Yes |
| 83 | Cog2 | 11495 | -0.478 | -0.1160 | Yes |
| 84 | Stx12 | 11662 | -0.511 | -0.1057 | Yes |
| 85 | Mon2 | 11747 | -0.534 | -0.0877 | Yes |
| 86 | Rps6ka3 | 11803 | -0.552 | -0.0665 | Yes |
| 87 | Pam | 11924 | -0.591 | -0.0487 | Yes |
| 88 | Vps4b | 12179 | -0.723 | -0.0357 | Yes |
| 89 | Clcn3 | 12338 | -1.088 | 0.0020 | Yes |

