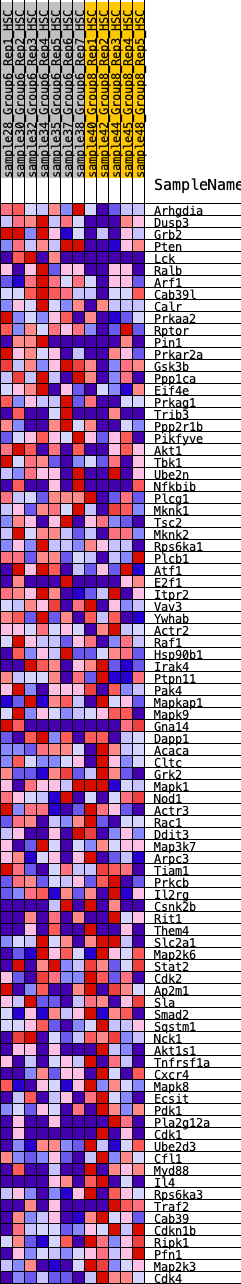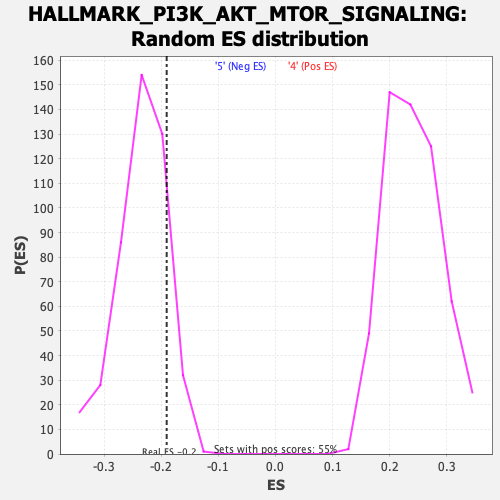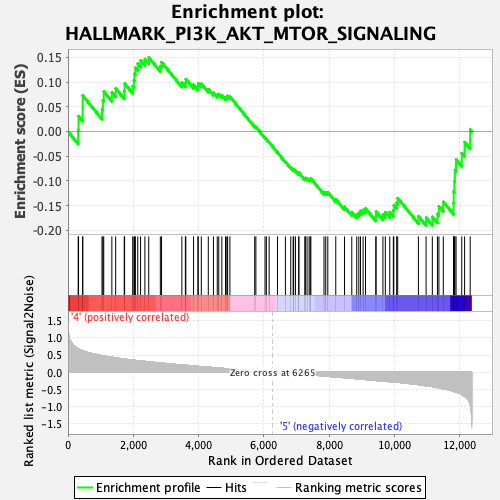
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.19050065 |
| Normalized Enrichment Score (NES) | -0.81760633 |
| Nominal p-value | 0.8504464 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Arhgdia | 311 | 0.689 | 0.0034 | No |
| 2 | Dusp3 | 323 | 0.683 | 0.0310 | No |
| 3 | Grb2 | 450 | 0.624 | 0.0467 | No |
| 4 | Pten | 451 | 0.623 | 0.0727 | No |
| 5 | Lck | 1041 | 0.487 | 0.0451 | No |
| 6 | Ralb | 1067 | 0.482 | 0.0632 | No |
| 7 | Arf1 | 1094 | 0.477 | 0.0810 | No |
| 8 | Cab39l | 1343 | 0.438 | 0.0790 | No |
| 9 | Calr | 1459 | 0.420 | 0.0871 | No |
| 10 | Prkaa2 | 1721 | 0.386 | 0.0820 | No |
| 11 | Rptor | 1733 | 0.385 | 0.0971 | No |
| 12 | Pin1 | 1981 | 0.354 | 0.0918 | No |
| 13 | Prkar2a | 2018 | 0.351 | 0.1035 | No |
| 14 | Gsk3b | 2036 | 0.350 | 0.1167 | No |
| 15 | Ppp1ca | 2063 | 0.346 | 0.1290 | No |
| 16 | Eif4e | 2136 | 0.339 | 0.1373 | No |
| 17 | Prkag1 | 2222 | 0.330 | 0.1441 | No |
| 18 | Trib3 | 2354 | 0.318 | 0.1467 | No |
| 19 | Ppp2r1b | 2472 | 0.305 | 0.1499 | No |
| 20 | Pikfyve | 2827 | 0.270 | 0.1323 | No |
| 21 | Akt1 | 2864 | 0.265 | 0.1405 | No |
| 22 | Tbk1 | 3486 | 0.215 | 0.0988 | No |
| 23 | Ube2n | 3593 | 0.205 | 0.0988 | No |
| 24 | Nfkbib | 3608 | 0.204 | 0.1061 | No |
| 25 | Plcg1 | 3844 | 0.185 | 0.0947 | No |
| 26 | Mknk1 | 3978 | 0.173 | 0.0910 | No |
| 27 | Tsc2 | 3986 | 0.172 | 0.0976 | No |
| 28 | Mknk2 | 4081 | 0.164 | 0.0968 | No |
| 29 | Rps6ka1 | 4295 | 0.149 | 0.0857 | No |
| 30 | Plcb1 | 4452 | 0.136 | 0.0787 | No |
| 31 | Atf1 | 4570 | 0.128 | 0.0745 | No |
| 32 | E2f1 | 4616 | 0.124 | 0.0760 | No |
| 33 | Itpr2 | 4710 | 0.116 | 0.0732 | No |
| 34 | Vav3 | 4823 | 0.107 | 0.0686 | No |
| 35 | Ywhab | 4862 | 0.105 | 0.0699 | No |
| 36 | Actr2 | 4887 | 0.102 | 0.0722 | No |
| 37 | Raf1 | 4957 | 0.098 | 0.0706 | No |
| 38 | Hsp90b1 | 5716 | 0.041 | 0.0106 | No |
| 39 | Irak4 | 5752 | 0.038 | 0.0093 | No |
| 40 | Ptpn11 | 6032 | 0.018 | -0.0127 | No |
| 41 | Pak4 | 6073 | 0.014 | -0.0154 | No |
| 42 | Mapkap1 | 6158 | 0.006 | -0.0220 | No |
| 43 | Mapk9 | 6414 | -0.005 | -0.0425 | No |
| 44 | Gna14 | 6656 | -0.021 | -0.0613 | No |
| 45 | Dapp1 | 6820 | -0.034 | -0.0732 | No |
| 46 | Acaca | 6893 | -0.039 | -0.0774 | No |
| 47 | Cltc | 6908 | -0.040 | -0.0769 | No |
| 48 | Grk2 | 6963 | -0.045 | -0.0794 | No |
| 49 | Mapk1 | 7066 | -0.052 | -0.0855 | No |
| 50 | Nod1 | 7067 | -0.052 | -0.0834 | No |
| 51 | Actr3 | 7251 | -0.066 | -0.0955 | No |
| 52 | Rac1 | 7266 | -0.067 | -0.0939 | No |
| 53 | Ddit3 | 7319 | -0.070 | -0.0952 | No |
| 54 | Map3k7 | 7382 | -0.075 | -0.0971 | No |
| 55 | Arpc3 | 7414 | -0.077 | -0.0965 | No |
| 56 | Tiam1 | 7436 | -0.078 | -0.0949 | No |
| 57 | Prkcb | 7838 | -0.106 | -0.1231 | No |
| 58 | Il2rg | 7891 | -0.111 | -0.1228 | No |
| 59 | Csnk2b | 7954 | -0.117 | -0.1229 | No |
| 60 | Rit1 | 8198 | -0.136 | -0.1371 | No |
| 61 | Them4 | 8466 | -0.156 | -0.1523 | No |
| 62 | Slc2a1 | 8688 | -0.173 | -0.1631 | No |
| 63 | Map2k6 | 8840 | -0.186 | -0.1676 | No |
| 64 | Stat2 | 8904 | -0.192 | -0.1648 | No |
| 65 | Cdk2 | 8956 | -0.196 | -0.1607 | No |
| 66 | Ap2m1 | 9035 | -0.204 | -0.1586 | No |
| 67 | Sla | 9109 | -0.211 | -0.1557 | No |
| 68 | Smad2 | 9425 | -0.239 | -0.1715 | No |
| 69 | Sqstm1 | 9431 | -0.239 | -0.1619 | No |
| 70 | Nck1 | 9642 | -0.255 | -0.1684 | No |
| 71 | Akt1s1 | 9715 | -0.262 | -0.1633 | No |
| 72 | Tnfrsf1a | 9851 | -0.272 | -0.1630 | No |
| 73 | Cxcr4 | 9960 | -0.281 | -0.1600 | No |
| 74 | Mapk8 | 9975 | -0.282 | -0.1494 | No |
| 75 | Ecsit | 10062 | -0.288 | -0.1444 | No |
| 76 | Pdk1 | 10094 | -0.291 | -0.1348 | No |
| 77 | Pla2g12a | 10728 | -0.358 | -0.1714 | No |
| 78 | Cdk1 | 10963 | -0.390 | -0.1742 | Yes |
| 79 | Ube2d3 | 11154 | -0.414 | -0.1725 | Yes |
| 80 | Cfl1 | 11311 | -0.445 | -0.1666 | Yes |
| 81 | Myd88 | 11356 | -0.455 | -0.1512 | Yes |
| 82 | Il4 | 11490 | -0.477 | -0.1421 | Yes |
| 83 | Rps6ka3 | 11803 | -0.552 | -0.1445 | Yes |
| 84 | Traf2 | 11808 | -0.553 | -0.1218 | Yes |
| 85 | Cab39 | 11828 | -0.560 | -0.1000 | Yes |
| 86 | Cdkn1b | 11845 | -0.564 | -0.0778 | Yes |
| 87 | Ripk1 | 11881 | -0.578 | -0.0565 | Yes |
| 88 | Pfn1 | 12055 | -0.646 | -0.0436 | Yes |
| 89 | Map2k3 | 12142 | -0.699 | -0.0215 | Yes |
| 90 | Cdk4 | 12313 | -0.945 | 0.0041 | Yes |