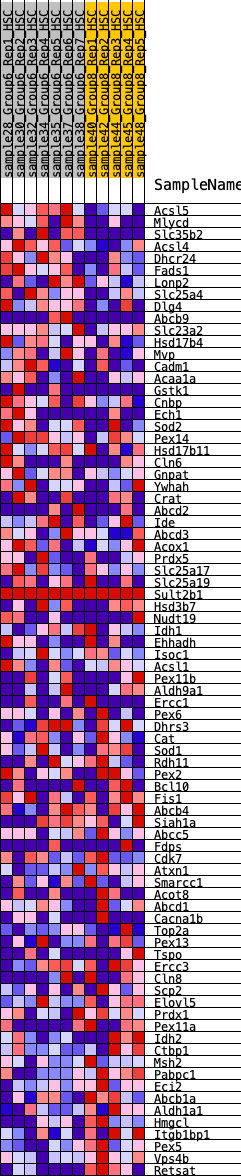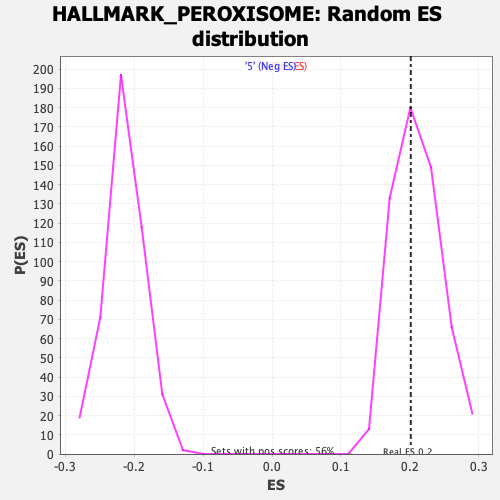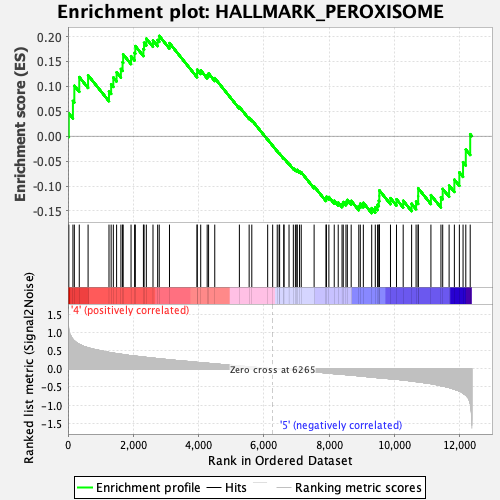
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.20132226 |
| Normalized Enrichment Score (NES) | 0.95408785 |
| Nominal p-value | 0.5996441 |
| FDR q-value | 0.95091826 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acsl5 | 25 | 1.073 | 0.0454 | Yes |
| 2 | Mlycd | 151 | 0.805 | 0.0709 | Yes |
| 3 | Slc35b2 | 194 | 0.762 | 0.1012 | Yes |
| 4 | Acsl4 | 345 | 0.670 | 0.1186 | Yes |
| 5 | Dhcr24 | 615 | 0.575 | 0.1222 | Yes |
| 6 | Fads1 | 1255 | 0.449 | 0.0900 | Yes |
| 7 | Lonp2 | 1321 | 0.440 | 0.1042 | Yes |
| 8 | Slc25a4 | 1388 | 0.430 | 0.1179 | Yes |
| 9 | Dlg4 | 1488 | 0.414 | 0.1281 | Yes |
| 10 | Abcb9 | 1620 | 0.398 | 0.1351 | Yes |
| 11 | Slc23a2 | 1671 | 0.391 | 0.1483 | Yes |
| 12 | Hsd17b4 | 1688 | 0.390 | 0.1642 | Yes |
| 13 | Mvp | 1930 | 0.360 | 0.1606 | Yes |
| 14 | Cadm1 | 2039 | 0.349 | 0.1672 | Yes |
| 15 | Acaa1a | 2061 | 0.347 | 0.1808 | Yes |
| 16 | Gstk1 | 2309 | 0.322 | 0.1750 | Yes |
| 17 | Cnbp | 2329 | 0.321 | 0.1876 | Yes |
| 18 | Ech1 | 2398 | 0.311 | 0.1959 | Yes |
| 19 | Sod2 | 2602 | 0.293 | 0.1923 | Yes |
| 20 | Pex14 | 2745 | 0.278 | 0.1930 | Yes |
| 21 | Hsd17b11 | 2793 | 0.274 | 0.2013 | Yes |
| 22 | Cln6 | 3107 | 0.248 | 0.1868 | No |
| 23 | Gnpat | 3952 | 0.175 | 0.1259 | No |
| 24 | Ywhah | 3954 | 0.175 | 0.1335 | No |
| 25 | Crat | 4063 | 0.166 | 0.1321 | No |
| 26 | Abcd2 | 4261 | 0.153 | 0.1228 | No |
| 27 | Ide | 4304 | 0.148 | 0.1259 | No |
| 28 | Abcd3 | 4493 | 0.133 | 0.1165 | No |
| 29 | Acox1 | 5247 | 0.078 | 0.0587 | No |
| 30 | Prdx5 | 5546 | 0.053 | 0.0368 | No |
| 31 | Slc25a17 | 5627 | 0.047 | 0.0323 | No |
| 32 | Slc25a19 | 6113 | 0.010 | -0.0067 | No |
| 33 | Sult2b1 | 6266 | 0.000 | -0.0191 | No |
| 34 | Hsd3b7 | 6406 | -0.004 | -0.0302 | No |
| 35 | Nudt19 | 6462 | -0.008 | -0.0343 | No |
| 36 | Idh1 | 6479 | -0.010 | -0.0352 | No |
| 37 | Ehhadh | 6607 | -0.018 | -0.0448 | No |
| 38 | Isoc1 | 6613 | -0.019 | -0.0444 | No |
| 39 | Acsl1 | 6767 | -0.029 | -0.0555 | No |
| 40 | Pex11b | 6899 | -0.039 | -0.0645 | No |
| 41 | Aldh9a1 | 6962 | -0.045 | -0.0675 | No |
| 42 | Ercc1 | 7007 | -0.048 | -0.0690 | No |
| 43 | Pex6 | 7017 | -0.049 | -0.0676 | No |
| 44 | Dhrs3 | 7089 | -0.054 | -0.0710 | No |
| 45 | Cat | 7141 | -0.058 | -0.0725 | No |
| 46 | Sod1 | 7535 | -0.085 | -0.1008 | No |
| 47 | Rdh11 | 7897 | -0.111 | -0.1252 | No |
| 48 | Pex2 | 7913 | -0.113 | -0.1214 | No |
| 49 | Bcl10 | 7987 | -0.119 | -0.1221 | No |
| 50 | Fis1 | 8151 | -0.133 | -0.1295 | No |
| 51 | Abcb4 | 8273 | -0.141 | -0.1331 | No |
| 52 | Siah1a | 8390 | -0.150 | -0.1359 | No |
| 53 | Abcc5 | 8427 | -0.153 | -0.1321 | No |
| 54 | Fdps | 8510 | -0.159 | -0.1317 | No |
| 55 | Cdk7 | 8552 | -0.162 | -0.1279 | No |
| 56 | Atxn1 | 8670 | -0.172 | -0.1298 | No |
| 57 | Smarcc1 | 8903 | -0.192 | -0.1402 | No |
| 58 | Acot8 | 8949 | -0.195 | -0.1352 | No |
| 59 | Abcd1 | 9042 | -0.204 | -0.1337 | No |
| 60 | Cacna1b | 9298 | -0.228 | -0.1444 | No |
| 61 | Top2a | 9408 | -0.237 | -0.1428 | No |
| 62 | Pex13 | 9478 | -0.244 | -0.1376 | No |
| 63 | Tspo | 9516 | -0.248 | -0.1296 | No |
| 64 | Ercc3 | 9531 | -0.249 | -0.1198 | No |
| 65 | Cln8 | 9532 | -0.249 | -0.1088 | No |
| 66 | Scp2 | 9873 | -0.273 | -0.1244 | No |
| 67 | Elovl5 | 10057 | -0.288 | -0.1265 | No |
| 68 | Prdx1 | 10262 | -0.308 | -0.1295 | No |
| 69 | Pex11a | 10520 | -0.336 | -0.1355 | No |
| 70 | Idh2 | 10657 | -0.351 | -0.1311 | No |
| 71 | Ctbp1 | 10720 | -0.357 | -0.1204 | No |
| 72 | Msh2 | 10722 | -0.357 | -0.1046 | No |
| 73 | Pabpc1 | 11111 | -0.407 | -0.1182 | No |
| 74 | Eci2 | 11418 | -0.468 | -0.1225 | No |
| 75 | Abcb1a | 11472 | -0.474 | -0.1058 | No |
| 76 | Aldh1a1 | 11668 | -0.513 | -0.0990 | No |
| 77 | Hmgcl | 11829 | -0.561 | -0.0872 | No |
| 78 | Itgb1bp1 | 11982 | -0.609 | -0.0726 | No |
| 79 | Pex5 | 12095 | -0.669 | -0.0521 | No |
| 80 | Vps4b | 12179 | -0.723 | -0.0269 | No |
| 81 | Retsat | 12314 | -0.945 | 0.0040 | No |