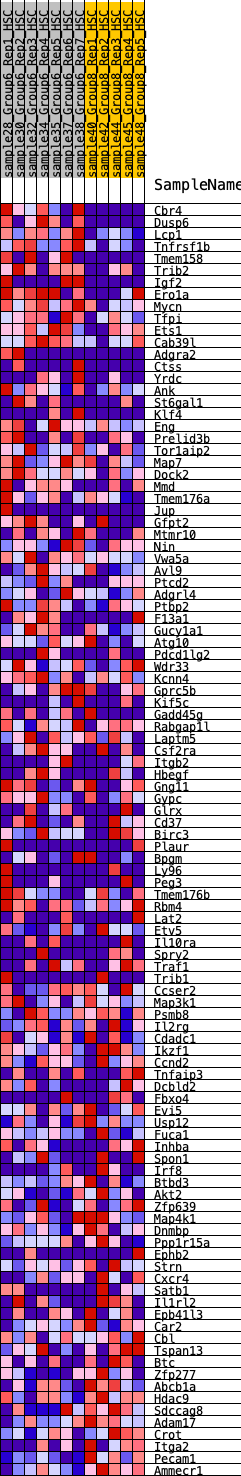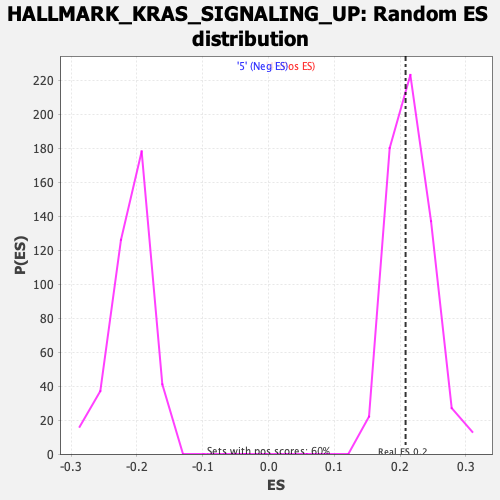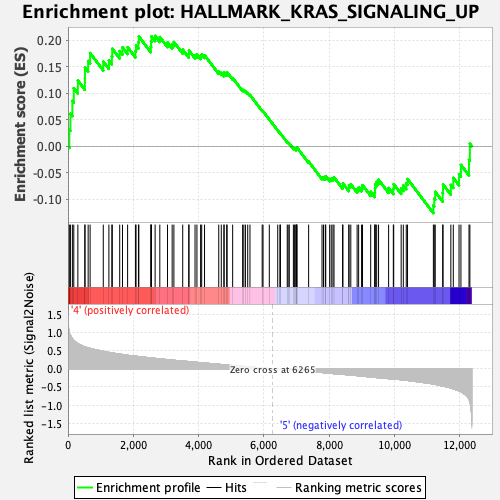
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.2083232 |
| Normalized Enrichment Score (NES) | 0.96630335 |
| Nominal p-value | 0.55481726 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cbr4 | 41 | 1.001 | 0.0318 | Yes |
| 2 | Dusp6 | 74 | 0.920 | 0.0615 | Yes |
| 3 | Lcp1 | 134 | 0.826 | 0.0857 | Yes |
| 4 | Tnfrsf1b | 177 | 0.779 | 0.1096 | Yes |
| 5 | Tmem158 | 302 | 0.696 | 0.1240 | Yes |
| 6 | Trib2 | 517 | 0.603 | 0.1277 | Yes |
| 7 | Igf2 | 519 | 0.601 | 0.1487 | Yes |
| 8 | Ero1a | 617 | 0.574 | 0.1610 | Yes |
| 9 | Mycn | 677 | 0.560 | 0.1758 | Yes |
| 10 | Tfpi | 1080 | 0.479 | 0.1599 | Yes |
| 11 | Ets1 | 1252 | 0.450 | 0.1617 | Yes |
| 12 | Cab39l | 1343 | 0.438 | 0.1697 | Yes |
| 13 | Adgra2 | 1355 | 0.436 | 0.1842 | Yes |
| 14 | Ctss | 1583 | 0.401 | 0.1798 | Yes |
| 15 | Yrdc | 1668 | 0.392 | 0.1867 | Yes |
| 16 | Ank | 1825 | 0.372 | 0.1870 | Yes |
| 17 | St6gal1 | 2064 | 0.346 | 0.1797 | Yes |
| 18 | Klf4 | 2083 | 0.343 | 0.1903 | Yes |
| 19 | Eng | 2157 | 0.338 | 0.1962 | Yes |
| 20 | Prelid3b | 2169 | 0.336 | 0.2072 | Yes |
| 21 | Tor1aip2 | 2535 | 0.299 | 0.1879 | Yes |
| 22 | Map7 | 2545 | 0.298 | 0.1976 | Yes |
| 23 | Dock2 | 2556 | 0.298 | 0.2073 | Yes |
| 24 | Mmd | 2667 | 0.286 | 0.2083 | Yes |
| 25 | Tmem176a | 2811 | 0.272 | 0.2062 | No |
| 26 | Jup | 3047 | 0.252 | 0.1959 | No |
| 27 | Gfpt2 | 3189 | 0.240 | 0.1928 | No |
| 28 | Mtmr10 | 3242 | 0.234 | 0.1968 | No |
| 29 | Nin | 3512 | 0.212 | 0.1823 | No |
| 30 | Vwa5a | 3697 | 0.199 | 0.1743 | No |
| 31 | Avl9 | 3704 | 0.198 | 0.1807 | No |
| 32 | Ptcd2 | 3889 | 0.181 | 0.1721 | No |
| 33 | Adgrl4 | 3947 | 0.176 | 0.1736 | No |
| 34 | Ptbp2 | 4060 | 0.166 | 0.1703 | No |
| 35 | F13a1 | 4093 | 0.163 | 0.1734 | No |
| 36 | Gucy1a1 | 4180 | 0.155 | 0.1719 | No |
| 37 | Atg10 | 4615 | 0.124 | 0.1408 | No |
| 38 | Pdcd1lg2 | 4693 | 0.117 | 0.1386 | No |
| 39 | Wdr33 | 4775 | 0.111 | 0.1359 | No |
| 40 | Kcnn4 | 4778 | 0.111 | 0.1397 | No |
| 41 | Gprc5b | 4860 | 0.105 | 0.1367 | No |
| 42 | Kif5c | 4870 | 0.104 | 0.1397 | No |
| 43 | Gadd45g | 5041 | 0.090 | 0.1290 | No |
| 44 | Rabgap1l | 5346 | 0.069 | 0.1066 | No |
| 45 | Laptm5 | 5383 | 0.066 | 0.1060 | No |
| 46 | Csf2ra | 5432 | 0.063 | 0.1042 | No |
| 47 | Itgb2 | 5502 | 0.057 | 0.1006 | No |
| 48 | Hbegf | 5571 | 0.052 | 0.0969 | No |
| 49 | Gng11 | 5945 | 0.024 | 0.0673 | No |
| 50 | Gypc | 5971 | 0.022 | 0.0660 | No |
| 51 | Glrx | 6163 | 0.006 | 0.0506 | No |
| 52 | Cd37 | 6421 | -0.006 | 0.0299 | No |
| 53 | Birc3 | 6487 | -0.010 | 0.0249 | No |
| 54 | Plaur | 6504 | -0.011 | 0.0240 | No |
| 55 | Bpgm | 6712 | -0.026 | 0.0080 | No |
| 56 | Ly96 | 6739 | -0.027 | 0.0069 | No |
| 57 | Peg3 | 6775 | -0.030 | 0.0050 | No |
| 58 | Tmem176b | 6900 | -0.039 | -0.0037 | No |
| 59 | Rbm4 | 6930 | -0.042 | -0.0046 | No |
| 60 | Lat2 | 6957 | -0.044 | -0.0051 | No |
| 61 | Etv5 | 6989 | -0.047 | -0.0060 | No |
| 62 | Il10ra | 6994 | -0.047 | -0.0047 | No |
| 63 | Spry2 | 6998 | -0.047 | -0.0033 | No |
| 64 | Traf1 | 7014 | -0.048 | -0.0028 | No |
| 65 | Trib1 | 7367 | -0.074 | -0.0289 | No |
| 66 | Ccser2 | 7777 | -0.102 | -0.0587 | No |
| 67 | Map3k1 | 7823 | -0.106 | -0.0587 | No |
| 68 | Psmb8 | 7878 | -0.110 | -0.0592 | No |
| 69 | Il2rg | 7891 | -0.111 | -0.0563 | No |
| 70 | Cdadc1 | 8011 | -0.122 | -0.0617 | No |
| 71 | Ikzf1 | 8059 | -0.126 | -0.0612 | No |
| 72 | Ccnd2 | 8111 | -0.130 | -0.0607 | No |
| 73 | Tnfaip3 | 8144 | -0.132 | -0.0587 | No |
| 74 | Dcbld2 | 8406 | -0.152 | -0.0747 | No |
| 75 | Fbxo4 | 8417 | -0.153 | -0.0701 | No |
| 76 | Evi5 | 8598 | -0.166 | -0.0790 | No |
| 77 | Usp12 | 8602 | -0.167 | -0.0734 | No |
| 78 | Fuca1 | 8658 | -0.171 | -0.0718 | No |
| 79 | Inhba | 8852 | -0.187 | -0.0810 | No |
| 80 | Spon1 | 8898 | -0.191 | -0.0779 | No |
| 81 | Irf8 | 8988 | -0.199 | -0.0782 | No |
| 82 | Btbd3 | 9017 | -0.203 | -0.0734 | No |
| 83 | Akt2 | 9267 | -0.224 | -0.0858 | No |
| 84 | Zfp639 | 9392 | -0.235 | -0.0877 | No |
| 85 | Map4k1 | 9399 | -0.235 | -0.0799 | No |
| 86 | Dnmbp | 9404 | -0.236 | -0.0719 | No |
| 87 | Ppp1r15a | 9450 | -0.241 | -0.0672 | No |
| 88 | Ephb2 | 9508 | -0.247 | -0.0631 | No |
| 89 | Strn | 9816 | -0.269 | -0.0787 | No |
| 90 | Cxcr4 | 9960 | -0.281 | -0.0805 | No |
| 91 | Satb1 | 9972 | -0.282 | -0.0715 | No |
| 92 | Il1rl2 | 10200 | -0.301 | -0.0795 | No |
| 93 | Epb41l3 | 10266 | -0.309 | -0.0739 | No |
| 94 | Car2 | 10356 | -0.318 | -0.0700 | No |
| 95 | Cbl | 10395 | -0.322 | -0.0618 | No |
| 96 | Tspan13 | 11186 | -0.420 | -0.1115 | No |
| 97 | Btc | 11214 | -0.425 | -0.0988 | No |
| 98 | Zfp277 | 11243 | -0.429 | -0.0860 | No |
| 99 | Abcb1a | 11472 | -0.474 | -0.0880 | No |
| 100 | Hdac9 | 11482 | -0.475 | -0.0720 | No |
| 101 | Sdccag8 | 11721 | -0.527 | -0.0729 | No |
| 102 | Adam17 | 11796 | -0.550 | -0.0596 | No |
| 103 | Crot | 11972 | -0.607 | -0.0526 | No |
| 104 | Itga2 | 12030 | -0.628 | -0.0352 | No |
| 105 | Pecam1 | 12277 | -0.848 | -0.0255 | No |
| 106 | Ammecr1 | 12304 | -0.922 | 0.0048 | No |