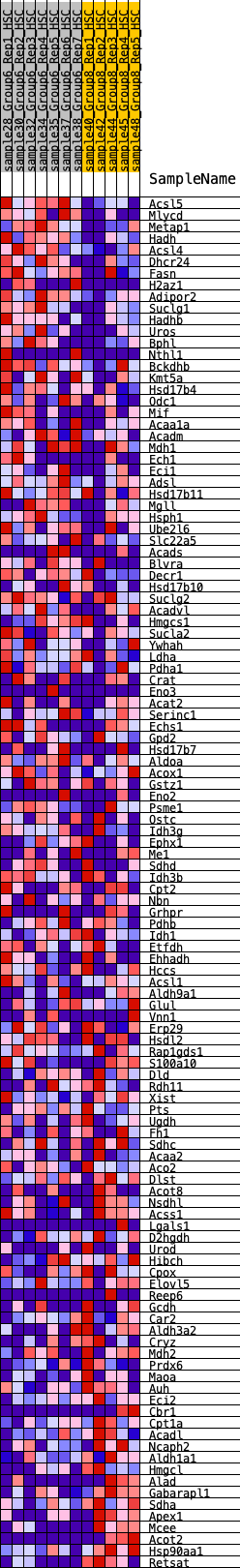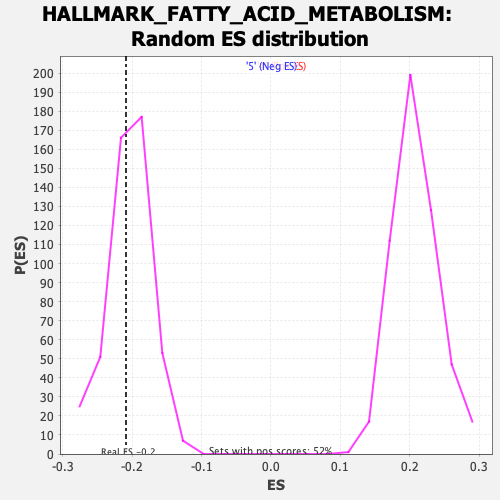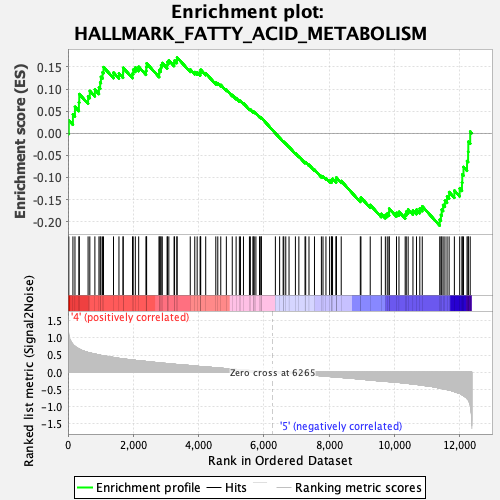
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.20898558 |
| Normalized Enrichment Score (NES) | -1.026429 |
| Nominal p-value | 0.3862213 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acsl5 | 25 | 1.073 | 0.0294 | No |
| 2 | Mlycd | 151 | 0.805 | 0.0428 | No |
| 3 | Metap1 | 213 | 0.752 | 0.0599 | No |
| 4 | Hadh | 334 | 0.675 | 0.0699 | No |
| 5 | Acsl4 | 345 | 0.670 | 0.0887 | No |
| 6 | Dhcr24 | 615 | 0.575 | 0.0836 | No |
| 7 | Fasn | 668 | 0.563 | 0.0959 | No |
| 8 | H2az1 | 823 | 0.529 | 0.0988 | No |
| 9 | Adipor2 | 948 | 0.504 | 0.1035 | No |
| 10 | Suclg1 | 985 | 0.497 | 0.1151 | No |
| 11 | Hadhb | 1008 | 0.490 | 0.1277 | No |
| 12 | Uros | 1058 | 0.485 | 0.1379 | No |
| 13 | Bphl | 1087 | 0.479 | 0.1496 | No |
| 14 | Nthl1 | 1393 | 0.429 | 0.1373 | No |
| 15 | Bckdhb | 1560 | 0.403 | 0.1356 | No |
| 16 | Kmt5a | 1686 | 0.390 | 0.1368 | No |
| 17 | Hsd17b4 | 1688 | 0.390 | 0.1481 | No |
| 18 | Odc1 | 1975 | 0.355 | 0.1352 | No |
| 19 | Mif | 1996 | 0.353 | 0.1439 | No |
| 20 | Acaa1a | 2061 | 0.347 | 0.1489 | No |
| 21 | Acadm | 2164 | 0.337 | 0.1504 | No |
| 22 | Mdh1 | 2389 | 0.313 | 0.1413 | No |
| 23 | Ech1 | 2398 | 0.311 | 0.1498 | No |
| 24 | Eci1 | 2407 | 0.311 | 0.1582 | No |
| 25 | Adsl | 2791 | 0.274 | 0.1350 | No |
| 26 | Hsd17b11 | 2793 | 0.274 | 0.1429 | No |
| 27 | Mgll | 2840 | 0.268 | 0.1470 | No |
| 28 | Hsph1 | 2849 | 0.268 | 0.1542 | No |
| 29 | Ube2l6 | 2886 | 0.263 | 0.1590 | No |
| 30 | Slc22a5 | 3037 | 0.253 | 0.1542 | No |
| 31 | Acads | 3042 | 0.253 | 0.1613 | No |
| 32 | Blvra | 3087 | 0.250 | 0.1650 | No |
| 33 | Decr1 | 3248 | 0.234 | 0.1588 | No |
| 34 | Hsd17b10 | 3262 | 0.232 | 0.1645 | No |
| 35 | Suclg2 | 3334 | 0.225 | 0.1653 | No |
| 36 | Acadvl | 3339 | 0.225 | 0.1716 | No |
| 37 | Hmgcs1 | 3743 | 0.193 | 0.1444 | No |
| 38 | Sucla2 | 3881 | 0.182 | 0.1385 | No |
| 39 | Ywhah | 3954 | 0.175 | 0.1378 | No |
| 40 | Ldha | 4046 | 0.167 | 0.1352 | No |
| 41 | Pdha1 | 4055 | 0.166 | 0.1395 | No |
| 42 | Crat | 4063 | 0.166 | 0.1437 | No |
| 43 | Eno3 | 4216 | 0.155 | 0.1359 | No |
| 44 | Acat2 | 4525 | 0.130 | 0.1145 | No |
| 45 | Serinc1 | 4586 | 0.126 | 0.1133 | No |
| 46 | Echs1 | 4677 | 0.118 | 0.1094 | No |
| 47 | Gpd2 | 4848 | 0.105 | 0.0986 | No |
| 48 | Hsd17b7 | 5029 | 0.091 | 0.0866 | No |
| 49 | Aldoa | 5147 | 0.084 | 0.0795 | No |
| 50 | Acox1 | 5247 | 0.078 | 0.0737 | No |
| 51 | Gstz1 | 5280 | 0.074 | 0.0733 | No |
| 52 | Eno2 | 5372 | 0.067 | 0.0678 | No |
| 53 | Psme1 | 5555 | 0.053 | 0.0545 | No |
| 54 | Ostc | 5583 | 0.051 | 0.0538 | No |
| 55 | Idh3g | 5667 | 0.044 | 0.0483 | No |
| 56 | Ephx1 | 5669 | 0.044 | 0.0495 | No |
| 57 | Me1 | 5702 | 0.041 | 0.0481 | No |
| 58 | Sdhd | 5753 | 0.038 | 0.0451 | No |
| 59 | Idh3b | 5863 | 0.030 | 0.0371 | No |
| 60 | Cpt2 | 5881 | 0.028 | 0.0365 | No |
| 61 | Nbn | 5916 | 0.026 | 0.0345 | No |
| 62 | Grhpr | 5918 | 0.026 | 0.0352 | No |
| 63 | Pdhb | 6349 | -0.000 | 0.0001 | No |
| 64 | Idh1 | 6479 | -0.010 | -0.0102 | No |
| 65 | Etfdh | 6587 | -0.017 | -0.0184 | No |
| 66 | Ehhadh | 6607 | -0.018 | -0.0194 | No |
| 67 | Hccs | 6667 | -0.022 | -0.0236 | No |
| 68 | Acsl1 | 6767 | -0.029 | -0.0308 | No |
| 69 | Aldh9a1 | 6962 | -0.045 | -0.0454 | No |
| 70 | Glul | 7068 | -0.052 | -0.0524 | No |
| 71 | Vnn1 | 7257 | -0.066 | -0.0658 | No |
| 72 | Erp29 | 7277 | -0.067 | -0.0654 | No |
| 73 | Hsdl2 | 7378 | -0.075 | -0.0714 | No |
| 74 | Rap1gds1 | 7546 | -0.086 | -0.0825 | No |
| 75 | S100a10 | 7758 | -0.101 | -0.0968 | No |
| 76 | Dld | 7807 | -0.104 | -0.0976 | No |
| 77 | Rdh11 | 7897 | -0.111 | -0.1016 | No |
| 78 | Xist | 8010 | -0.122 | -0.1072 | No |
| 79 | Pts | 8069 | -0.127 | -0.1082 | No |
| 80 | Ugdh | 8075 | -0.128 | -0.1049 | No |
| 81 | Fh1 | 8099 | -0.129 | -0.1030 | No |
| 82 | Sdhc | 8209 | -0.137 | -0.1079 | No |
| 83 | Acaa2 | 8212 | -0.137 | -0.1040 | No |
| 84 | Aco2 | 8214 | -0.137 | -0.1001 | No |
| 85 | Dlst | 8365 | -0.148 | -0.1080 | No |
| 86 | Acot8 | 8949 | -0.195 | -0.1498 | No |
| 87 | Nsdhl | 8968 | -0.198 | -0.1455 | No |
| 88 | Acss1 | 9253 | -0.223 | -0.1622 | No |
| 89 | Lgals1 | 9592 | -0.252 | -0.1824 | No |
| 90 | D2hgdh | 9716 | -0.262 | -0.1848 | No |
| 91 | Urod | 9774 | -0.267 | -0.1816 | No |
| 92 | Hibch | 9831 | -0.270 | -0.1783 | No |
| 93 | Cpox | 9833 | -0.270 | -0.1704 | No |
| 94 | Elovl5 | 10057 | -0.288 | -0.1802 | No |
| 95 | Reep6 | 10133 | -0.295 | -0.1777 | No |
| 96 | Gcdh | 10323 | -0.314 | -0.1839 | No |
| 97 | Car2 | 10356 | -0.318 | -0.1772 | No |
| 98 | Aldh3a2 | 10414 | -0.323 | -0.1724 | No |
| 99 | Cryz | 10561 | -0.341 | -0.1743 | No |
| 100 | Mdh2 | 10670 | -0.352 | -0.1728 | No |
| 101 | Prdx6 | 10771 | -0.364 | -0.1703 | No |
| 102 | Maoa | 10846 | -0.374 | -0.1654 | No |
| 103 | Auh | 11381 | -0.458 | -0.1955 | Yes |
| 104 | Eci2 | 11418 | -0.468 | -0.1848 | Yes |
| 105 | Cbr1 | 11442 | -0.471 | -0.1728 | Yes |
| 106 | Cpt1a | 11484 | -0.476 | -0.1622 | Yes |
| 107 | Acadl | 11540 | -0.486 | -0.1525 | Yes |
| 108 | Ncaph2 | 11605 | -0.501 | -0.1430 | Yes |
| 109 | Aldh1a1 | 11668 | -0.513 | -0.1330 | Yes |
| 110 | Hmgcl | 11829 | -0.561 | -0.1297 | Yes |
| 111 | Alad | 11992 | -0.612 | -0.1250 | Yes |
| 112 | Gabarapl1 | 12062 | -0.648 | -0.1116 | Yes |
| 113 | Sdha | 12071 | -0.654 | -0.0931 | Yes |
| 114 | Apex1 | 12109 | -0.675 | -0.0763 | Yes |
| 115 | Mcee | 12217 | -0.760 | -0.0627 | Yes |
| 116 | Acot2 | 12250 | -0.795 | -0.0420 | Yes |
| 117 | Hsp90aa1 | 12256 | -0.798 | -0.0191 | Yes |
| 118 | Retsat | 12314 | -0.945 | 0.0040 | Yes |