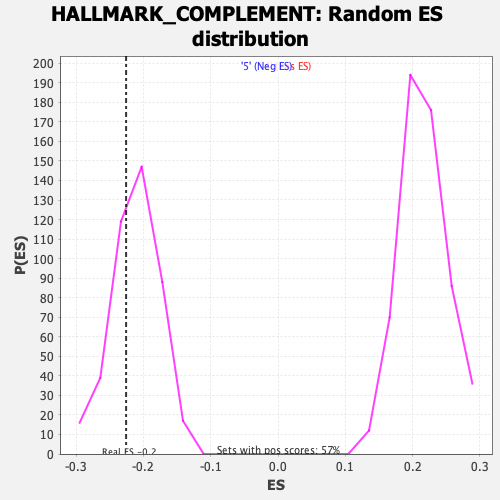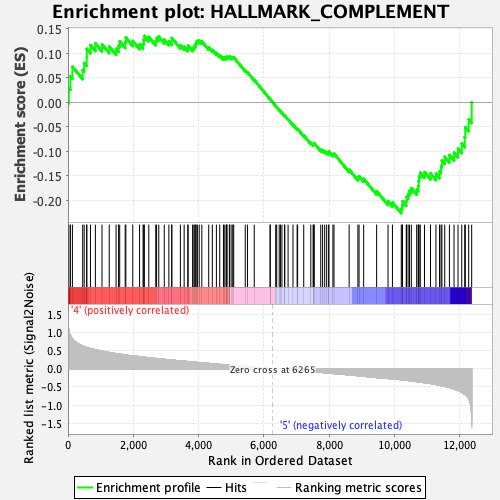
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.22580096 |
| Normalized Enrichment Score (NES) | -1.0632248 |
| Nominal p-value | 0.31455398 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lap3 | 27 | 1.065 | 0.0296 | No |
| 2 | Dusp6 | 74 | 0.920 | 0.0534 | No |
| 3 | Gnb2 | 135 | 0.826 | 0.0731 | No |
| 4 | Grb2 | 450 | 0.624 | 0.0661 | No |
| 5 | Gnai2 | 495 | 0.609 | 0.0808 | No |
| 6 | Dock9 | 573 | 0.588 | 0.0920 | No |
| 7 | Cblb | 575 | 0.587 | 0.1095 | No |
| 8 | Itgam | 687 | 0.558 | 0.1171 | No |
| 9 | Fyn | 838 | 0.527 | 0.1206 | No |
| 10 | Lck | 1041 | 0.487 | 0.1187 | No |
| 11 | Cd46 | 1262 | 0.448 | 0.1141 | No |
| 12 | Atox1 | 1472 | 0.417 | 0.1095 | No |
| 13 | Kynu | 1548 | 0.406 | 0.1155 | No |
| 14 | Ctss | 1583 | 0.401 | 0.1248 | No |
| 15 | Gng2 | 1751 | 0.382 | 0.1225 | No |
| 16 | Csrp1 | 1771 | 0.380 | 0.1323 | No |
| 17 | Lyn | 1983 | 0.354 | 0.1257 | No |
| 18 | Dgkg | 2191 | 0.334 | 0.1188 | No |
| 19 | F5 | 2298 | 0.323 | 0.1198 | No |
| 20 | Hspa5 | 2319 | 0.322 | 0.1278 | No |
| 21 | Col4a2 | 2337 | 0.320 | 0.1359 | No |
| 22 | Usp15 | 2474 | 0.305 | 0.1340 | No |
| 23 | Adam9 | 2687 | 0.284 | 0.1251 | No |
| 24 | Pclo | 2712 | 0.281 | 0.1316 | No |
| 25 | Lta4h | 2779 | 0.275 | 0.1344 | No |
| 26 | Pdp1 | 2947 | 0.259 | 0.1285 | No |
| 27 | Dyrk2 | 3088 | 0.250 | 0.1245 | No |
| 28 | Casp4 | 3176 | 0.241 | 0.1246 | No |
| 29 | Cpq | 3178 | 0.241 | 0.1317 | No |
| 30 | Prss36 | 3440 | 0.219 | 0.1170 | No |
| 31 | Spock2 | 3556 | 0.208 | 0.1138 | No |
| 32 | Gca | 3669 | 0.201 | 0.1107 | No |
| 33 | Kif2a | 3676 | 0.200 | 0.1162 | No |
| 34 | S100a13 | 3814 | 0.187 | 0.1106 | No |
| 35 | Ehd1 | 3854 | 0.184 | 0.1129 | No |
| 36 | Xpnpep1 | 3882 | 0.182 | 0.1161 | No |
| 37 | Plek | 3911 | 0.179 | 0.1192 | No |
| 38 | Msrb1 | 3921 | 0.178 | 0.1238 | No |
| 39 | Plscr1 | 3962 | 0.174 | 0.1257 | No |
| 40 | Rhog | 4017 | 0.169 | 0.1264 | No |
| 41 | Akap10 | 4097 | 0.163 | 0.1248 | No |
| 42 | Dock10 | 4308 | 0.148 | 0.1121 | No |
| 43 | Psmb9 | 4418 | 0.139 | 0.1073 | No |
| 44 | Gnai3 | 4543 | 0.129 | 0.1010 | No |
| 45 | Zeb1 | 4646 | 0.121 | 0.0963 | No |
| 46 | Pla2g4a | 4759 | 0.112 | 0.0905 | No |
| 47 | Lrp1 | 4782 | 0.110 | 0.0920 | No |
| 48 | Ctsl | 4839 | 0.106 | 0.0906 | No |
| 49 | Gpd2 | 4848 | 0.105 | 0.0931 | No |
| 50 | Brpf3 | 4871 | 0.104 | 0.0944 | No |
| 51 | Prep | 4939 | 0.100 | 0.0919 | No |
| 52 | Raf1 | 4957 | 0.098 | 0.0935 | No |
| 53 | Lipa | 5018 | 0.092 | 0.0913 | No |
| 54 | Stx4a | 5048 | 0.090 | 0.0916 | No |
| 55 | Calm3 | 5073 | 0.088 | 0.0923 | No |
| 56 | Timp2 | 5426 | 0.063 | 0.0654 | No |
| 57 | Pim1 | 5494 | 0.058 | 0.0617 | No |
| 58 | Me1 | 5702 | 0.041 | 0.0460 | No |
| 59 | Casp7 | 6188 | 0.004 | 0.0065 | No |
| 60 | Casp3 | 6192 | 0.004 | 0.0064 | No |
| 61 | Dock4 | 6359 | -0.001 | -0.0071 | No |
| 62 | Ppp2cb | 6389 | -0.003 | -0.0094 | No |
| 63 | Mmp14 | 6466 | -0.009 | -0.0153 | No |
| 64 | Plaur | 6504 | -0.011 | -0.0180 | No |
| 65 | Dusp5 | 6550 | -0.014 | -0.0213 | No |
| 66 | Usp16 | 6631 | -0.020 | -0.0272 | No |
| 67 | C2 | 6636 | -0.020 | -0.0269 | No |
| 68 | Rce1 | 6738 | -0.027 | -0.0344 | No |
| 69 | Lamp2 | 6888 | -0.038 | -0.0454 | No |
| 70 | Sirt6 | 7015 | -0.048 | -0.0542 | No |
| 71 | Irf1 | 7030 | -0.049 | -0.0539 | No |
| 72 | Fdx1 | 7217 | -0.064 | -0.0672 | No |
| 73 | Prkcd | 7433 | -0.078 | -0.0824 | No |
| 74 | Ctsh | 7504 | -0.084 | -0.0856 | No |
| 75 | Ctsc | 7519 | -0.084 | -0.0842 | No |
| 76 | Psen1 | 7540 | -0.086 | -0.0833 | No |
| 77 | Anxa5 | 7738 | -0.099 | -0.0964 | No |
| 78 | Was | 7795 | -0.103 | -0.0979 | No |
| 79 | Irf2 | 7859 | -0.108 | -0.0999 | No |
| 80 | Rbsn | 7919 | -0.114 | -0.1013 | No |
| 81 | Sh2b3 | 7982 | -0.119 | -0.1028 | No |
| 82 | Ctso | 7986 | -0.119 | -0.0995 | No |
| 83 | Gata3 | 8110 | -0.130 | -0.1056 | No |
| 84 | Tnfaip3 | 8144 | -0.132 | -0.1044 | No |
| 85 | Casp9 | 8607 | -0.167 | -0.1371 | No |
| 86 | Gnb4 | 8875 | -0.189 | -0.1533 | No |
| 87 | Usp8 | 8911 | -0.192 | -0.1504 | No |
| 88 | Dgkh | 9052 | -0.205 | -0.1557 | No |
| 89 | Clu | 9449 | -0.241 | -0.1808 | No |
| 90 | F8 | 9798 | -0.268 | -0.2012 | No |
| 91 | Dpp4 | 9933 | -0.279 | -0.2038 | No |
| 92 | Rabif | 10203 | -0.301 | -0.2168 | Yes |
| 93 | Kcnip3 | 10230 | -0.304 | -0.2098 | Yes |
| 94 | Pik3cg | 10242 | -0.306 | -0.2016 | Yes |
| 95 | Car2 | 10356 | -0.318 | -0.2013 | Yes |
| 96 | Ctsd | 10366 | -0.319 | -0.1925 | Yes |
| 97 | Ctsb | 10413 | -0.322 | -0.1867 | Yes |
| 98 | S100a9 | 10456 | -0.328 | -0.1803 | Yes |
| 99 | Irf7 | 10511 | -0.336 | -0.1747 | Yes |
| 100 | Jak2 | 10675 | -0.353 | -0.1774 | Yes |
| 101 | Vcpip1 | 10721 | -0.357 | -0.1704 | Yes |
| 102 | Casp1 | 10733 | -0.359 | -0.1606 | Yes |
| 103 | Rnf4 | 10748 | -0.361 | -0.1510 | Yes |
| 104 | Src | 10785 | -0.366 | -0.1430 | Yes |
| 105 | Prdm4 | 10914 | -0.382 | -0.1420 | Yes |
| 106 | Lcp2 | 11098 | -0.405 | -0.1448 | Yes |
| 107 | Gp9 | 11265 | -0.433 | -0.1454 | Yes |
| 108 | Pik3r5 | 11375 | -0.457 | -0.1407 | Yes |
| 109 | Prcp | 11421 | -0.468 | -0.1304 | Yes |
| 110 | Cr2 | 11447 | -0.471 | -0.1183 | Yes |
| 111 | Pdgfb | 11534 | -0.484 | -0.1109 | Yes |
| 112 | Maff | 11677 | -0.515 | -0.1071 | Yes |
| 113 | Cpm | 11819 | -0.557 | -0.1019 | Yes |
| 114 | Calm1 | 11941 | -0.595 | -0.0940 | Yes |
| 115 | Pfn1 | 12055 | -0.646 | -0.0839 | Yes |
| 116 | Pik3ca | 12147 | -0.700 | -0.0704 | Yes |
| 117 | Ppp4c | 12163 | -0.711 | -0.0504 | Yes |
| 118 | Gmfb | 12267 | -0.812 | -0.0345 | Yes |
| 119 | Usp14 | 12359 | -1.415 | 0.0003 | Yes |