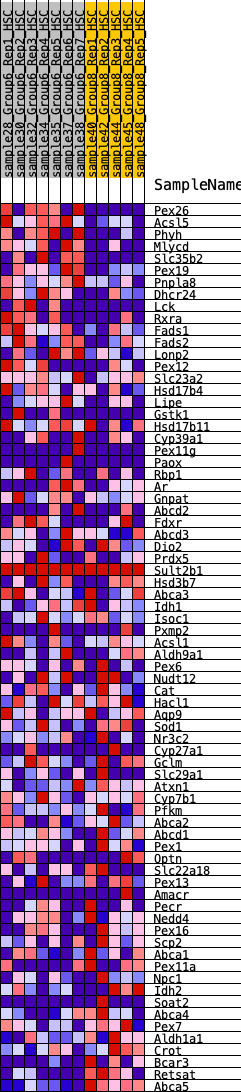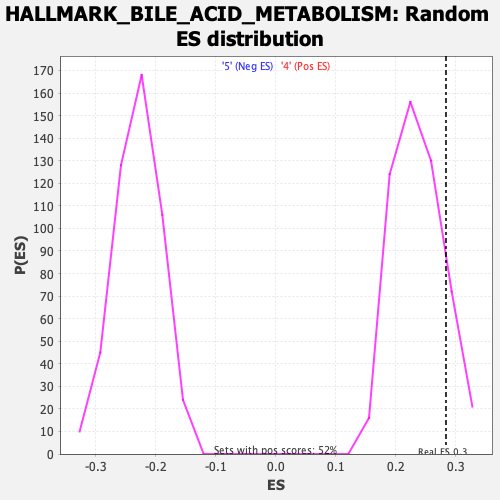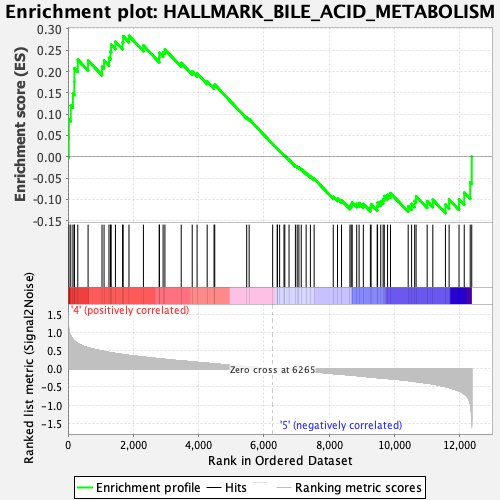
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.28353015 |
| Normalized Enrichment Score (NES) | 1.1987458 |
| Nominal p-value | 0.14643545 |
| FDR q-value | 0.47367194 |
| FWER p-Value | 0.925 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex26 | 22 | 1.107 | 0.0438 | Yes |
| 2 | Acsl5 | 25 | 1.073 | 0.0878 | Yes |
| 3 | Phyh | 85 | 0.905 | 0.1202 | Yes |
| 4 | Mlycd | 151 | 0.805 | 0.1481 | Yes |
| 5 | Slc35b2 | 194 | 0.762 | 0.1760 | Yes |
| 6 | Pex19 | 196 | 0.762 | 0.2073 | Yes |
| 7 | Pnpla8 | 300 | 0.696 | 0.2276 | Yes |
| 8 | Dhcr24 | 615 | 0.575 | 0.2257 | Yes |
| 9 | Lck | 1041 | 0.487 | 0.2112 | Yes |
| 10 | Rxra | 1104 | 0.476 | 0.2257 | Yes |
| 11 | Fads1 | 1255 | 0.449 | 0.2320 | Yes |
| 12 | Fads2 | 1301 | 0.443 | 0.2466 | Yes |
| 13 | Lonp2 | 1321 | 0.440 | 0.2631 | Yes |
| 14 | Pex12 | 1452 | 0.421 | 0.2698 | Yes |
| 15 | Slc23a2 | 1671 | 0.391 | 0.2682 | Yes |
| 16 | Hsd17b4 | 1688 | 0.390 | 0.2830 | Yes |
| 17 | Lipe | 1867 | 0.366 | 0.2835 | Yes |
| 18 | Gstk1 | 2309 | 0.322 | 0.2609 | No |
| 19 | Hsd17b11 | 2793 | 0.274 | 0.2329 | No |
| 20 | Cyp39a1 | 2798 | 0.273 | 0.2438 | No |
| 21 | Pex11g | 2911 | 0.262 | 0.2455 | No |
| 22 | Paox | 2967 | 0.257 | 0.2516 | No |
| 23 | Rbp1 | 3466 | 0.216 | 0.2200 | No |
| 24 | Ar | 3803 | 0.188 | 0.2004 | No |
| 25 | Gnpat | 3952 | 0.175 | 0.1955 | No |
| 26 | Abcd2 | 4261 | 0.153 | 0.1768 | No |
| 27 | Fdxr | 4471 | 0.134 | 0.1653 | No |
| 28 | Abcd3 | 4493 | 0.133 | 0.1691 | No |
| 29 | Dio2 | 5471 | 0.059 | 0.0920 | No |
| 30 | Prdx5 | 5546 | 0.053 | 0.0882 | No |
| 31 | Sult2b1 | 6266 | 0.000 | 0.0297 | No |
| 32 | Hsd3b7 | 6406 | -0.004 | 0.0186 | No |
| 33 | Abca3 | 6411 | -0.005 | 0.0184 | No |
| 34 | Idh1 | 6479 | -0.010 | 0.0134 | No |
| 35 | Isoc1 | 6613 | -0.019 | 0.0033 | No |
| 36 | Pxmp2 | 6638 | -0.020 | 0.0022 | No |
| 37 | Acsl1 | 6767 | -0.029 | -0.0070 | No |
| 38 | Aldh9a1 | 6962 | -0.045 | -0.0210 | No |
| 39 | Pex6 | 7017 | -0.049 | -0.0234 | No |
| 40 | Nudt12 | 7069 | -0.053 | -0.0253 | No |
| 41 | Cat | 7141 | -0.058 | -0.0287 | No |
| 42 | Hacl1 | 7291 | -0.068 | -0.0380 | No |
| 43 | Aqp9 | 7421 | -0.077 | -0.0454 | No |
| 44 | Sod1 | 7535 | -0.085 | -0.0510 | No |
| 45 | Nr3c2 | 8121 | -0.131 | -0.0932 | No |
| 46 | Cyp27a1 | 8251 | -0.140 | -0.0980 | No |
| 47 | Gclm | 8374 | -0.149 | -0.1018 | No |
| 48 | Slc29a1 | 8632 | -0.169 | -0.1157 | No |
| 49 | Atxn1 | 8670 | -0.172 | -0.1116 | No |
| 50 | Cyp7b1 | 8701 | -0.174 | -0.1069 | No |
| 51 | Pfkm | 8839 | -0.186 | -0.1104 | No |
| 52 | Abca2 | 8912 | -0.192 | -0.1084 | No |
| 53 | Abcd1 | 9042 | -0.204 | -0.1104 | No |
| 54 | Pex1 | 9258 | -0.223 | -0.1188 | No |
| 55 | Optn | 9279 | -0.225 | -0.1111 | No |
| 56 | Slc22a18 | 9466 | -0.243 | -0.1163 | No |
| 57 | Pex13 | 9478 | -0.244 | -0.1071 | No |
| 58 | Amacr | 9575 | -0.252 | -0.1046 | No |
| 59 | Pecr | 9648 | -0.256 | -0.0999 | No |
| 60 | Nedd4 | 9683 | -0.259 | -0.0920 | No |
| 61 | Pex16 | 9783 | -0.267 | -0.0890 | No |
| 62 | Scp2 | 9873 | -0.273 | -0.0850 | No |
| 63 | Abca1 | 10415 | -0.323 | -0.1158 | No |
| 64 | Pex11a | 10520 | -0.336 | -0.1104 | No |
| 65 | Npc1 | 10614 | -0.348 | -0.1036 | No |
| 66 | Idh2 | 10657 | -0.351 | -0.0926 | No |
| 67 | Soat2 | 10996 | -0.393 | -0.1039 | No |
| 68 | Abca4 | 11169 | -0.416 | -0.1008 | No |
| 69 | Pex7 | 11555 | -0.489 | -0.1120 | No |
| 70 | Aldh1a1 | 11668 | -0.513 | -0.1000 | No |
| 71 | Crot | 11972 | -0.607 | -0.0996 | No |
| 72 | Bcar3 | 12131 | -0.692 | -0.0840 | No |
| 73 | Retsat | 12314 | -0.945 | -0.0599 | No |
| 74 | Abca5 | 12361 | -1.550 | 0.0002 | No |