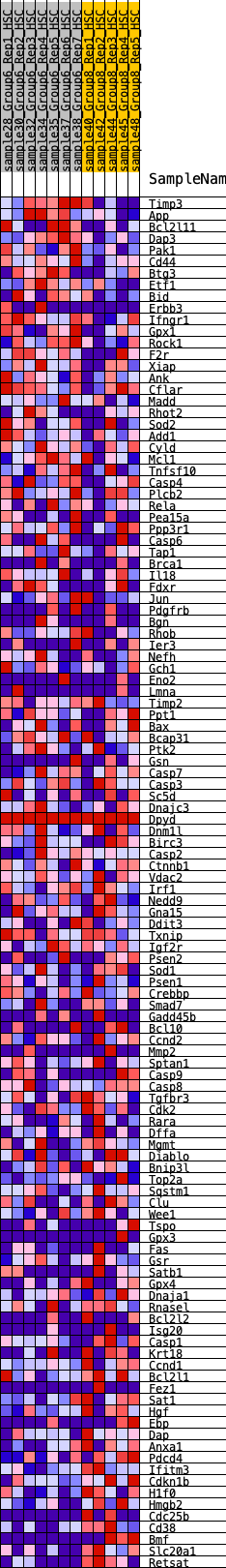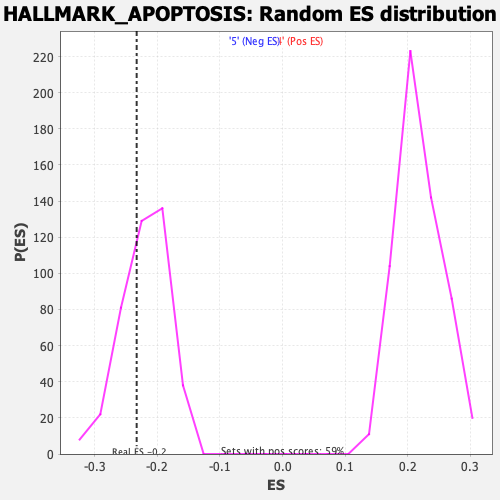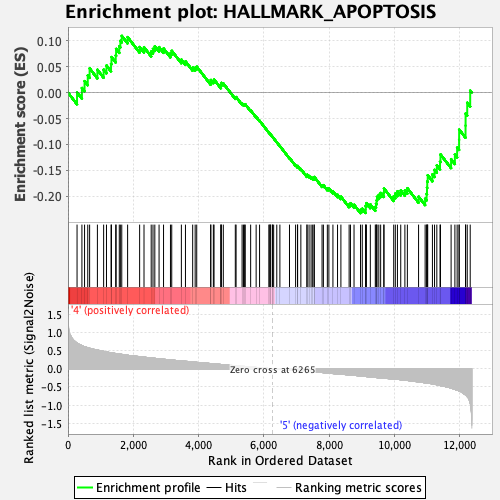
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group6_versus_Group8.HSC_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | HSC_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.23282067 |
| Normalized Enrichment Score (NES) | -1.0565375 |
| Nominal p-value | 0.34057972 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Timp3 | 277 | 0.707 | 0.0004 | No |
| 2 | App | 426 | 0.634 | 0.0089 | No |
| 3 | Bcl2l11 | 507 | 0.606 | 0.0222 | No |
| 4 | Dap3 | 605 | 0.577 | 0.0330 | No |
| 5 | Pak1 | 662 | 0.564 | 0.0468 | No |
| 6 | Cd44 | 900 | 0.515 | 0.0442 | No |
| 7 | Btg3 | 1091 | 0.478 | 0.0443 | No |
| 8 | Etf1 | 1178 | 0.464 | 0.0524 | No |
| 9 | Bid | 1318 | 0.441 | 0.0554 | No |
| 10 | Erbb3 | 1331 | 0.439 | 0.0687 | No |
| 11 | Ifngr1 | 1461 | 0.419 | 0.0718 | No |
| 12 | Gpx1 | 1473 | 0.417 | 0.0845 | No |
| 13 | Rock1 | 1570 | 0.402 | 0.0898 | No |
| 14 | F2r | 1603 | 0.399 | 0.1001 | No |
| 15 | Xiap | 1646 | 0.396 | 0.1096 | No |
| 16 | Ank | 1825 | 0.372 | 0.1072 | No |
| 17 | Cflar | 2193 | 0.334 | 0.0881 | No |
| 18 | Madd | 2327 | 0.321 | 0.0877 | No |
| 19 | Rhot2 | 2547 | 0.298 | 0.0795 | No |
| 20 | Sod2 | 2602 | 0.293 | 0.0847 | No |
| 21 | Add1 | 2658 | 0.287 | 0.0895 | No |
| 22 | Cyld | 2788 | 0.274 | 0.0879 | No |
| 23 | Mcl1 | 2925 | 0.261 | 0.0853 | No |
| 24 | Tnfsf10 | 3137 | 0.245 | 0.0760 | No |
| 25 | Casp4 | 3176 | 0.241 | 0.0808 | No |
| 26 | Plcb2 | 3471 | 0.215 | 0.0638 | No |
| 27 | Rela | 3596 | 0.205 | 0.0604 | No |
| 28 | Pea15a | 3817 | 0.187 | 0.0485 | No |
| 29 | Ppp3r1 | 3893 | 0.181 | 0.0482 | No |
| 30 | Casp6 | 3941 | 0.177 | 0.0501 | No |
| 31 | Tap1 | 4369 | 0.143 | 0.0199 | No |
| 32 | Brca1 | 4372 | 0.143 | 0.0244 | No |
| 33 | Il18 | 4450 | 0.137 | 0.0226 | No |
| 34 | Fdxr | 4471 | 0.134 | 0.0254 | No |
| 35 | Jun | 4681 | 0.118 | 0.0121 | No |
| 36 | Pdgfrb | 4682 | 0.118 | 0.0160 | No |
| 37 | Bgn | 4695 | 0.117 | 0.0188 | No |
| 38 | Rhob | 4755 | 0.112 | 0.0177 | No |
| 39 | Ier3 | 5122 | 0.086 | -0.0094 | No |
| 40 | Nefh | 5146 | 0.084 | -0.0086 | No |
| 41 | Gch1 | 5330 | 0.070 | -0.0212 | No |
| 42 | Eno2 | 5372 | 0.067 | -0.0224 | No |
| 43 | Lmna | 5407 | 0.064 | -0.0231 | No |
| 44 | Timp2 | 5426 | 0.063 | -0.0225 | No |
| 45 | Ppt1 | 5592 | 0.050 | -0.0343 | No |
| 46 | Bax | 5760 | 0.037 | -0.0468 | No |
| 47 | Bcap31 | 5865 | 0.030 | -0.0543 | No |
| 48 | Ptk2 | 6149 | 0.007 | -0.0772 | No |
| 49 | Gsn | 6181 | 0.005 | -0.0796 | No |
| 50 | Casp7 | 6188 | 0.004 | -0.0799 | No |
| 51 | Casp3 | 6192 | 0.004 | -0.0800 | No |
| 52 | Sc5d | 6256 | 0.000 | -0.0852 | No |
| 53 | Dnajc3 | 6258 | 0.000 | -0.0852 | No |
| 54 | Dpyd | 6296 | 0.000 | -0.0883 | No |
| 55 | Dnm1l | 6392 | -0.003 | -0.0959 | No |
| 56 | Birc3 | 6487 | -0.010 | -0.1033 | No |
| 57 | Casp2 | 6781 | -0.030 | -0.1262 | No |
| 58 | Ctnnb1 | 6968 | -0.045 | -0.1399 | No |
| 59 | Vdac2 | 7027 | -0.049 | -0.1431 | No |
| 60 | Irf1 | 7030 | -0.049 | -0.1416 | No |
| 61 | Nedd9 | 7129 | -0.057 | -0.1478 | No |
| 62 | Gna15 | 7315 | -0.070 | -0.1606 | No |
| 63 | Ddit3 | 7319 | -0.070 | -0.1585 | No |
| 64 | Txnip | 7380 | -0.075 | -0.1610 | No |
| 65 | Igf2r | 7442 | -0.078 | -0.1634 | No |
| 66 | Psen2 | 7491 | -0.083 | -0.1647 | No |
| 67 | Sod1 | 7535 | -0.085 | -0.1654 | No |
| 68 | Psen1 | 7540 | -0.086 | -0.1629 | No |
| 69 | Crebbp | 7778 | -0.102 | -0.1790 | No |
| 70 | Smad7 | 7819 | -0.105 | -0.1788 | No |
| 71 | Gadd45b | 7940 | -0.116 | -0.1848 | No |
| 72 | Bcl10 | 7987 | -0.119 | -0.1847 | No |
| 73 | Ccnd2 | 8111 | -0.130 | -0.1905 | No |
| 74 | Mmp2 | 8252 | -0.140 | -0.1974 | No |
| 75 | Sptan1 | 8355 | -0.148 | -0.2009 | No |
| 76 | Casp9 | 8607 | -0.167 | -0.2159 | No |
| 77 | Casp8 | 8648 | -0.171 | -0.2137 | No |
| 78 | Tgfbr3 | 8757 | -0.179 | -0.2167 | No |
| 79 | Cdk2 | 8956 | -0.196 | -0.2264 | Yes |
| 80 | Rara | 9009 | -0.201 | -0.2241 | Yes |
| 81 | Dffa | 9113 | -0.211 | -0.2256 | Yes |
| 82 | Mgmt | 9114 | -0.211 | -0.2188 | Yes |
| 83 | Diablo | 9136 | -0.213 | -0.2135 | Yes |
| 84 | Bnip3l | 9256 | -0.223 | -0.2160 | Yes |
| 85 | Top2a | 9408 | -0.237 | -0.2206 | Yes |
| 86 | Sqstm1 | 9431 | -0.239 | -0.2146 | Yes |
| 87 | Clu | 9449 | -0.241 | -0.2082 | Yes |
| 88 | Wee1 | 9460 | -0.242 | -0.2011 | Yes |
| 89 | Tspo | 9516 | -0.248 | -0.1976 | Yes |
| 90 | Gpx3 | 9572 | -0.252 | -0.1938 | Yes |
| 91 | Fas | 9669 | -0.258 | -0.1933 | Yes |
| 92 | Gsr | 9674 | -0.258 | -0.1852 | Yes |
| 93 | Satb1 | 9972 | -0.282 | -0.2003 | Yes |
| 94 | Gpx4 | 10021 | -0.287 | -0.1948 | Yes |
| 95 | Dnaja1 | 10086 | -0.290 | -0.1906 | Yes |
| 96 | Rnasel | 10187 | -0.300 | -0.1890 | Yes |
| 97 | Bcl2l2 | 10314 | -0.313 | -0.1891 | Yes |
| 98 | Isg20 | 10390 | -0.322 | -0.1848 | Yes |
| 99 | Casp1 | 10733 | -0.359 | -0.2010 | Yes |
| 100 | Krt18 | 10934 | -0.386 | -0.2048 | Yes |
| 101 | Ccnd1 | 10980 | -0.392 | -0.1957 | Yes |
| 102 | Bcl2l1 | 10991 | -0.393 | -0.1837 | Yes |
| 103 | Fez1 | 10999 | -0.393 | -0.1715 | Yes |
| 104 | Sat1 | 11013 | -0.394 | -0.1597 | Yes |
| 105 | Hgf | 11157 | -0.414 | -0.1579 | Yes |
| 106 | Ebp | 11222 | -0.426 | -0.1492 | Yes |
| 107 | Dap | 11292 | -0.440 | -0.1405 | Yes |
| 108 | Anxa1 | 11390 | -0.460 | -0.1335 | Yes |
| 109 | Pdcd4 | 11404 | -0.465 | -0.1194 | Yes |
| 110 | Ifitm3 | 11729 | -0.529 | -0.1286 | Yes |
| 111 | Cdkn1b | 11845 | -0.564 | -0.1197 | Yes |
| 112 | H1f0 | 11914 | -0.589 | -0.1060 | Yes |
| 113 | Hmgb2 | 11974 | -0.608 | -0.0910 | Yes |
| 114 | Cdc25b | 11976 | -0.609 | -0.0713 | Yes |
| 115 | Cd38 | 12171 | -0.719 | -0.0637 | Yes |
| 116 | Bmf | 12173 | -0.720 | -0.0404 | Yes |
| 117 | Slc20a1 | 12223 | -0.766 | -0.0194 | Yes |
| 118 | Retsat | 12314 | -0.945 | 0.0040 | Yes |