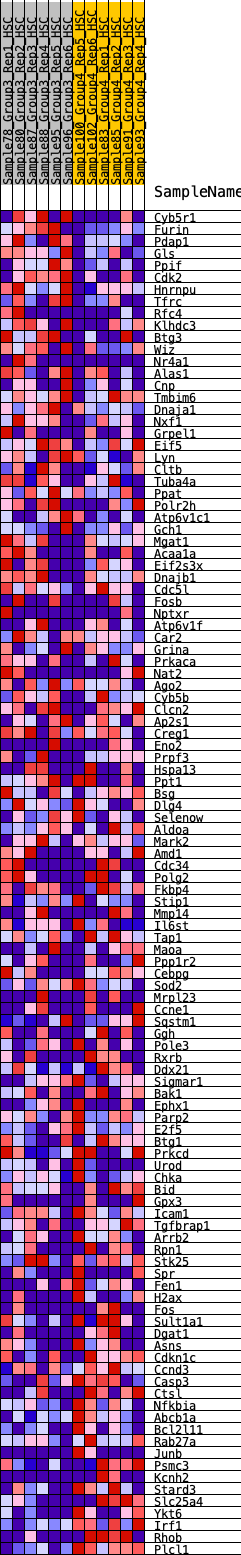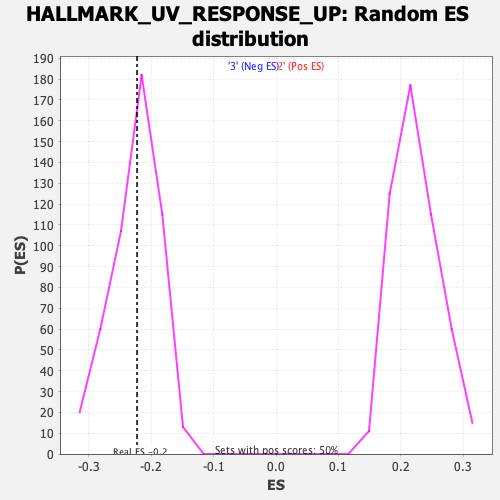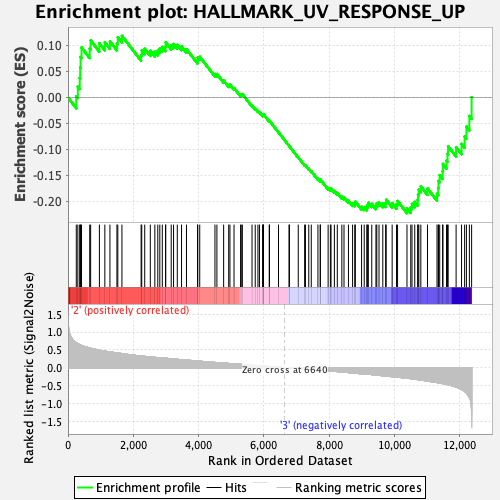
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.22252862 |
| Normalized Enrichment Score (NES) | -0.990241 |
| Nominal p-value | 0.45070422 |
| FDR q-value | 0.83026093 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyb5r1 | 254 | 0.699 | 0.0023 | No |
| 2 | Furin | 301 | 0.673 | 0.0207 | No |
| 3 | Pdap1 | 359 | 0.650 | 0.0375 | No |
| 4 | Gls | 374 | 0.644 | 0.0576 | No |
| 5 | Ppif | 386 | 0.637 | 0.0777 | No |
| 6 | Cdk2 | 414 | 0.625 | 0.0961 | No |
| 7 | Hnrnpu | 665 | 0.554 | 0.0940 | No |
| 8 | Tfrc | 693 | 0.546 | 0.1098 | No |
| 9 | Rfc4 | 961 | 0.492 | 0.1042 | No |
| 10 | Klhdc3 | 1128 | 0.464 | 0.1060 | No |
| 11 | Btg3 | 1284 | 0.443 | 0.1079 | No |
| 12 | Wiz | 1498 | 0.415 | 0.1042 | No |
| 13 | Nr4a1 | 1525 | 0.411 | 0.1156 | No |
| 14 | Alas1 | 1651 | 0.395 | 0.1184 | No |
| 15 | Cnp | 2238 | 0.327 | 0.0814 | No |
| 16 | Tmbim6 | 2258 | 0.325 | 0.0905 | No |
| 17 | Dnaja1 | 2349 | 0.317 | 0.0937 | No |
| 18 | Nxf1 | 2520 | 0.301 | 0.0897 | No |
| 19 | Grpel1 | 2659 | 0.289 | 0.0880 | No |
| 20 | Eif5 | 2749 | 0.284 | 0.0901 | No |
| 21 | Lyn | 2811 | 0.278 | 0.0943 | No |
| 22 | Cltb | 2887 | 0.273 | 0.0971 | No |
| 23 | Tuba4a | 2988 | 0.263 | 0.0976 | No |
| 24 | Ppat | 2991 | 0.262 | 0.1061 | No |
| 25 | Polr2h | 3160 | 0.250 | 0.1007 | No |
| 26 | Atp6v1c1 | 3232 | 0.244 | 0.1029 | No |
| 27 | Gch1 | 3346 | 0.234 | 0.1014 | No |
| 28 | Mgat1 | 3476 | 0.224 | 0.0982 | No |
| 29 | Acaa1a | 3627 | 0.209 | 0.0929 | No |
| 30 | Eif2s3x | 3966 | 0.187 | 0.0715 | No |
| 31 | Dnajb1 | 3973 | 0.187 | 0.0771 | No |
| 32 | Cdc5l | 4033 | 0.182 | 0.0783 | No |
| 33 | Fosb | 4499 | 0.147 | 0.0452 | No |
| 34 | Nptxr | 4559 | 0.143 | 0.0451 | No |
| 35 | Atp6v1f | 4765 | 0.128 | 0.0326 | No |
| 36 | Car2 | 4920 | 0.118 | 0.0240 | No |
| 37 | Grina | 4958 | 0.115 | 0.0247 | No |
| 38 | Prkaca | 5083 | 0.107 | 0.0181 | No |
| 39 | Nat2 | 5284 | 0.092 | 0.0049 | No |
| 40 | Ago2 | 5303 | 0.091 | 0.0064 | No |
| 41 | Cyb5b | 5338 | 0.089 | 0.0065 | No |
| 42 | Clcn2 | 5634 | 0.067 | -0.0153 | No |
| 43 | Ap2s1 | 5732 | 0.061 | -0.0213 | No |
| 44 | Creg1 | 5815 | 0.054 | -0.0262 | No |
| 45 | Eno2 | 5861 | 0.051 | -0.0282 | No |
| 46 | Prpf3 | 5957 | 0.045 | -0.0344 | No |
| 47 | Hspa13 | 5975 | 0.044 | -0.0344 | No |
| 48 | Ppt1 | 5985 | 0.043 | -0.0337 | No |
| 49 | Bsg | 5986 | 0.043 | -0.0323 | No |
| 50 | Dlg4 | 6164 | 0.033 | -0.0456 | No |
| 51 | Selenow | 6165 | 0.033 | -0.0445 | No |
| 52 | Aldoa | 6445 | 0.012 | -0.0669 | No |
| 53 | Mark2 | 6767 | -0.001 | -0.0931 | No |
| 54 | Amd1 | 6779 | -0.002 | -0.0939 | No |
| 55 | Cdc34 | 7049 | -0.021 | -0.1151 | No |
| 56 | Polg2 | 7245 | -0.035 | -0.1299 | No |
| 57 | Fkbp4 | 7270 | -0.037 | -0.1306 | No |
| 58 | Stip1 | 7371 | -0.043 | -0.1374 | No |
| 59 | Mmp14 | 7448 | -0.050 | -0.1419 | No |
| 60 | Il6st | 7652 | -0.064 | -0.1564 | No |
| 61 | Tap1 | 7723 | -0.069 | -0.1598 | No |
| 62 | Maoa | 7725 | -0.070 | -0.1576 | No |
| 63 | Ppp1r2 | 7964 | -0.088 | -0.1741 | No |
| 64 | Cebpg | 8033 | -0.094 | -0.1766 | No |
| 65 | Sod2 | 8052 | -0.096 | -0.1749 | No |
| 66 | Mrpl23 | 8155 | -0.103 | -0.1798 | No |
| 67 | Ccne1 | 8245 | -0.109 | -0.1835 | No |
| 68 | Sqstm1 | 8384 | -0.121 | -0.1908 | No |
| 69 | Ggh | 8450 | -0.127 | -0.1919 | No |
| 70 | Pole3 | 8586 | -0.136 | -0.1984 | No |
| 71 | Rxrb | 8713 | -0.148 | -0.2039 | No |
| 72 | Ddx21 | 8780 | -0.155 | -0.2041 | No |
| 73 | Sigmar1 | 8797 | -0.157 | -0.2002 | No |
| 74 | Bak1 | 8988 | -0.171 | -0.2101 | No |
| 75 | Ephx1 | 9069 | -0.176 | -0.2108 | No |
| 76 | Parp2 | 9148 | -0.184 | -0.2112 | No |
| 77 | E2f5 | 9166 | -0.186 | -0.2064 | No |
| 78 | Btg1 | 9195 | -0.188 | -0.2025 | No |
| 79 | Prkcd | 9300 | -0.194 | -0.2046 | No |
| 80 | Urod | 9428 | -0.205 | -0.2082 | No |
| 81 | Chka | 9454 | -0.208 | -0.2034 | No |
| 82 | Bid | 9522 | -0.216 | -0.2017 | No |
| 83 | Gpx3 | 9638 | -0.224 | -0.2037 | No |
| 84 | Icam1 | 9725 | -0.232 | -0.2031 | No |
| 85 | Tgfbrap1 | 9741 | -0.233 | -0.1967 | No |
| 86 | Arrb2 | 9924 | -0.253 | -0.2032 | No |
| 87 | Rpn1 | 10058 | -0.264 | -0.2053 | No |
| 88 | Stk25 | 10090 | -0.266 | -0.1991 | No |
| 89 | Spr | 10378 | -0.295 | -0.2128 | Yes |
| 90 | Fen1 | 10494 | -0.309 | -0.2120 | Yes |
| 91 | H2ax | 10532 | -0.314 | -0.2047 | Yes |
| 92 | Fos | 10616 | -0.322 | -0.2008 | Yes |
| 93 | Sult1a1 | 10706 | -0.335 | -0.1971 | Yes |
| 94 | Dgat1 | 10718 | -0.336 | -0.1869 | Yes |
| 95 | Asns | 10741 | -0.339 | -0.1775 | Yes |
| 96 | Cdkn1c | 10802 | -0.349 | -0.1709 | Yes |
| 97 | Ccnd3 | 11006 | -0.377 | -0.1750 | Yes |
| 98 | Casp3 | 11298 | -0.417 | -0.1850 | Yes |
| 99 | Ctsl | 11336 | -0.422 | -0.1741 | Yes |
| 100 | Nfkbia | 11343 | -0.424 | -0.1606 | Yes |
| 101 | Abcb1a | 11380 | -0.429 | -0.1494 | Yes |
| 102 | Bcl2l11 | 11470 | -0.445 | -0.1420 | Yes |
| 103 | Rab27a | 11478 | -0.447 | -0.1278 | Yes |
| 104 | Junb | 11589 | -0.467 | -0.1214 | Yes |
| 105 | Psmc3 | 11620 | -0.475 | -0.1082 | Yes |
| 106 | Kcnh2 | 11638 | -0.479 | -0.0938 | Yes |
| 107 | Stard3 | 11882 | -0.543 | -0.0957 | Yes |
| 108 | Slc25a4 | 12049 | -0.607 | -0.0892 | Yes |
| 109 | Ykt6 | 12143 | -0.662 | -0.0750 | Yes |
| 110 | Irf1 | 12198 | -0.709 | -0.0560 | Yes |
| 111 | Rhob | 12288 | -0.841 | -0.0356 | Yes |
| 112 | Plcl1 | 12357 | -1.262 | 0.0005 | Yes |