Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
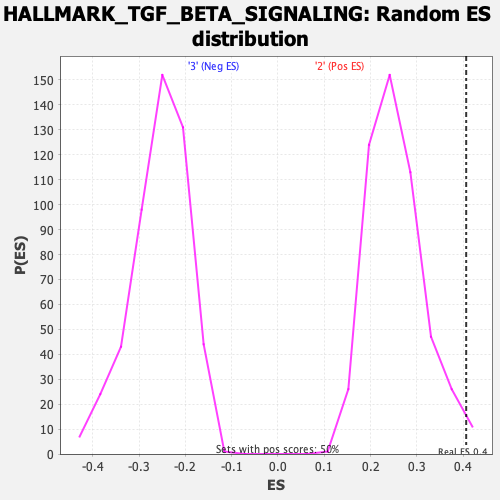
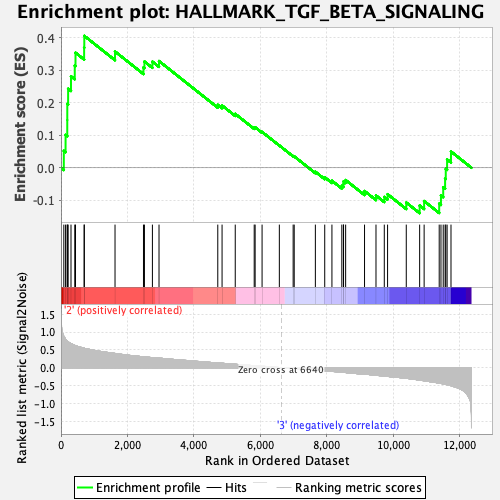
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.40625772 |
| Normalized Enrichment Score (NES) | 1.5935372 |
| Nominal p-value | 0.014 |
| FDR q-value | 0.15580703 |
| FWER p-Value | 0.13 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
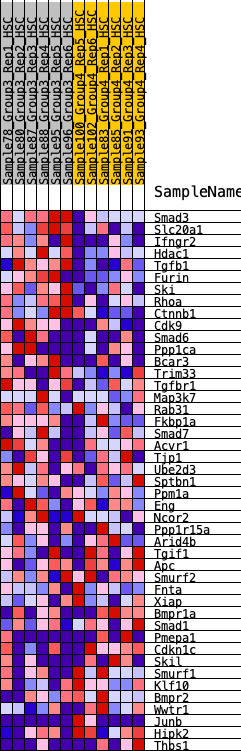
| 1 | Smad3 | 85 | 0.879 | 0.0523 | Yes |
| 2 | Slc20a1 | 138 | 0.790 | 0.1013 | Yes |
| 3 | Ifngr2 | 189 | 0.740 | 0.1471 | Yes |
| 4 | Hdac1 | 190 | 0.740 | 0.1969 | Yes |
| 5 | Tgfb1 | 215 | 0.719 | 0.2434 | Yes |
| 6 | Furin | 301 | 0.673 | 0.2818 | Yes |
| 7 | Ski | 415 | 0.625 | 0.3147 | Yes |
| 8 | Rhoa | 437 | 0.615 | 0.3543 | Yes |
| 9 | Ctnnb1 | 697 | 0.545 | 0.3700 | Yes |
| 10 | Cdk9 | 702 | 0.543 | 0.4063 | Yes |
| 11 | Smad6 | 1627 | 0.399 | 0.3581 | No |
| 12 | Ppp1ca | 2484 | 0.305 | 0.3091 | No |
| 13 | Bcar3 | 2513 | 0.302 | 0.3272 | No |
| 14 | Trim33 | 2753 | 0.283 | 0.3269 | No |
| 15 | Tgfbr1 | 2952 | 0.265 | 0.3287 | No |
| 16 | Map3k7 | 4722 | 0.132 | 0.1939 | No |
| 17 | Rab31 | 4851 | 0.123 | 0.1918 | No |
| 18 | Fkbp1a | 5248 | 0.095 | 0.1661 | No |
| 19 | Smad7 | 5819 | 0.054 | 0.1234 | No |
| 20 | Acvr1 | 5847 | 0.051 | 0.1247 | No |
| 21 | Tjp1 | 6056 | 0.040 | 0.1105 | No |
| 22 | Ube2d3 | 6576 | 0.005 | 0.0687 | No |
| 23 | Sptbn1 | 6992 | -0.018 | 0.0362 | No |
| 24 | Ppm1a | 7024 | -0.020 | 0.0351 | No |
| 25 | Eng | 7661 | -0.064 | -0.0122 | No |
| 26 | Ncor2 | 7943 | -0.086 | -0.0292 | No |
| 27 | Ppp1r15a | 8160 | -0.103 | -0.0398 | No |
| 28 | Arid4b | 8458 | -0.127 | -0.0554 | No |
| 29 | Tgif1 | 8507 | -0.130 | -0.0505 | No |
| 30 | Apc | 8510 | -0.131 | -0.0419 | No |
| 31 | Smurf2 | 8573 | -0.135 | -0.0378 | No |
| 32 | Fnta | 9142 | -0.183 | -0.0716 | No |
| 33 | Xiap | 9487 | -0.212 | -0.0853 | No |
| 34 | Bmpr1a | 9737 | -0.233 | -0.0898 | No |
| 35 | Smad1 | 9836 | -0.243 | -0.0814 | No |
| 36 | Pmepa1 | 10399 | -0.297 | -0.1070 | No |
| 37 | Cdkn1c | 10802 | -0.349 | -0.1162 | No |
| 38 | Skil | 10937 | -0.365 | -0.1025 | No |
| 39 | Smurf1 | 11391 | -0.429 | -0.1103 | No |
| 40 | Klf10 | 11444 | -0.439 | -0.0850 | No |
| 41 | Bmpr2 | 11512 | -0.453 | -0.0599 | No |
| 42 | Wwtr1 | 11569 | -0.463 | -0.0333 | No |
| 43 | Junb | 11589 | -0.467 | -0.0034 | No |
| 44 | Hipk2 | 11629 | -0.478 | 0.0256 | No |
| 45 | Thbs1 | 11746 | -0.504 | 0.0501 | No |