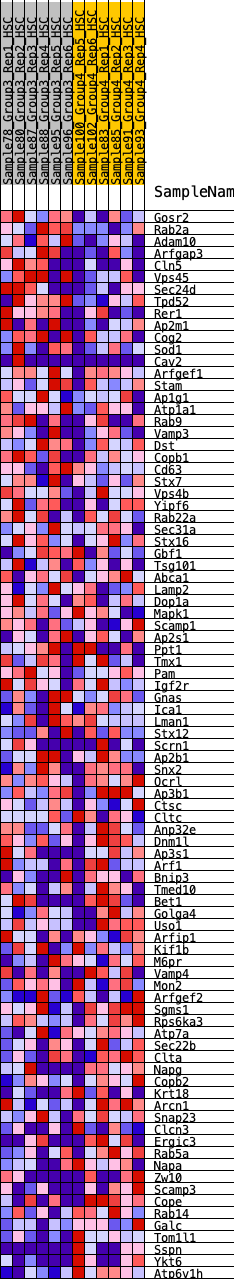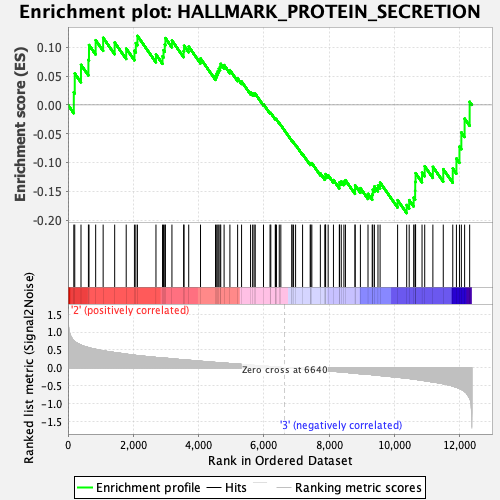
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.18803842 |
| Normalized Enrichment Score (NES) | -0.7971601 |
| Nominal p-value | 0.78642714 |
| FDR q-value | 0.985024 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gosr2 | 178 | 0.753 | 0.0219 | No |
| 2 | Rab2a | 207 | 0.723 | 0.0546 | No |
| 3 | Adam10 | 398 | 0.632 | 0.0697 | No |
| 4 | Arfgap3 | 625 | 0.563 | 0.0784 | No |
| 5 | Cln5 | 643 | 0.559 | 0.1041 | No |
| 6 | Vps45 | 847 | 0.514 | 0.1124 | No |
| 7 | Sec24d | 1076 | 0.472 | 0.1166 | No |
| 8 | Tpd52 | 1429 | 0.422 | 0.1084 | No |
| 9 | Rer1 | 1782 | 0.378 | 0.0979 | No |
| 10 | Ap2m1 | 2034 | 0.348 | 0.0943 | No |
| 11 | Cog2 | 2077 | 0.344 | 0.1075 | No |
| 12 | Sod1 | 2126 | 0.338 | 0.1199 | No |
| 13 | Cav2 | 2693 | 0.286 | 0.0877 | No |
| 14 | Arfgef1 | 2894 | 0.272 | 0.0845 | No |
| 15 | Stam | 2925 | 0.269 | 0.0951 | No |
| 16 | Ap1g1 | 2963 | 0.265 | 0.1049 | No |
| 17 | Atp1a1 | 2982 | 0.263 | 0.1161 | No |
| 18 | Rab9 | 3182 | 0.249 | 0.1119 | No |
| 19 | Vamp3 | 3540 | 0.218 | 0.0934 | No |
| 20 | Dst | 3553 | 0.217 | 0.1029 | No |
| 21 | Copb1 | 3696 | 0.207 | 0.1013 | No |
| 22 | Cd63 | 4057 | 0.180 | 0.0807 | No |
| 23 | Stx7 | 4516 | 0.146 | 0.0504 | No |
| 24 | Vps4b | 4553 | 0.143 | 0.0544 | No |
| 25 | Yipf6 | 4587 | 0.142 | 0.0586 | No |
| 26 | Rab22a | 4618 | 0.139 | 0.0629 | No |
| 27 | Sec31a | 4655 | 0.136 | 0.0665 | No |
| 28 | Stx16 | 4676 | 0.134 | 0.0714 | No |
| 29 | Gbf1 | 4779 | 0.127 | 0.0692 | No |
| 30 | Tsg101 | 4957 | 0.115 | 0.0604 | No |
| 31 | Abca1 | 5193 | 0.099 | 0.0460 | No |
| 32 | Lamp2 | 5314 | 0.090 | 0.0406 | No |
| 33 | Dop1a | 5589 | 0.071 | 0.0217 | No |
| 34 | Mapk1 | 5653 | 0.065 | 0.0197 | No |
| 35 | Scamp1 | 5696 | 0.063 | 0.0194 | No |
| 36 | Ap2s1 | 5732 | 0.061 | 0.0194 | No |
| 37 | Ppt1 | 5985 | 0.043 | 0.0010 | No |
| 38 | Tmx1 | 6186 | 0.032 | -0.0137 | No |
| 39 | Pam | 6209 | 0.030 | -0.0141 | No |
| 40 | Igf2r | 6346 | 0.019 | -0.0242 | No |
| 41 | Gnas | 6353 | 0.019 | -0.0238 | No |
| 42 | Ica1 | 6382 | 0.017 | -0.0253 | No |
| 43 | Lman1 | 6468 | 0.011 | -0.0317 | No |
| 44 | Stx12 | 6516 | 0.008 | -0.0351 | No |
| 45 | Scrn1 | 6854 | -0.008 | -0.0622 | No |
| 46 | Ap2b1 | 6858 | -0.008 | -0.0621 | No |
| 47 | Snx2 | 6906 | -0.012 | -0.0654 | No |
| 48 | Ocrl | 6968 | -0.016 | -0.0696 | No |
| 49 | Ap3b1 | 7182 | -0.029 | -0.0855 | No |
| 50 | Ctsc | 7415 | -0.047 | -0.1021 | No |
| 51 | Cltc | 7433 | -0.049 | -0.1012 | No |
| 52 | Anp32e | 7465 | -0.051 | -0.1012 | No |
| 53 | Dnm1l | 7726 | -0.070 | -0.1190 | No |
| 54 | Ap3s1 | 7863 | -0.079 | -0.1263 | No |
| 55 | Arf1 | 7885 | -0.081 | -0.1241 | No |
| 56 | Bnip3 | 7886 | -0.081 | -0.1201 | No |
| 57 | Tmed10 | 7965 | -0.088 | -0.1222 | No |
| 58 | Bet1 | 8127 | -0.100 | -0.1305 | No |
| 59 | Golga4 | 8307 | -0.115 | -0.1395 | No |
| 60 | Uso1 | 8313 | -0.115 | -0.1344 | No |
| 61 | Arfip1 | 8366 | -0.119 | -0.1329 | No |
| 62 | Kif1b | 8447 | -0.126 | -0.1333 | No |
| 63 | M6pr | 8494 | -0.129 | -0.1308 | No |
| 64 | Vamp4 | 8787 | -0.156 | -0.1470 | No |
| 65 | Mon2 | 8790 | -0.157 | -0.1396 | No |
| 66 | Arfgef2 | 8952 | -0.168 | -0.1446 | No |
| 67 | Sgms1 | 9185 | -0.187 | -0.1545 | No |
| 68 | Rps6ka3 | 9314 | -0.195 | -0.1554 | No |
| 69 | Atp7a | 9331 | -0.196 | -0.1473 | No |
| 70 | Sec22b | 9381 | -0.200 | -0.1416 | No |
| 71 | Clta | 9490 | -0.212 | -0.1401 | No |
| 72 | Napg | 9555 | -0.218 | -0.1348 | No |
| 73 | Copb2 | 10091 | -0.266 | -0.1655 | No |
| 74 | Krt18 | 10369 | -0.294 | -0.1738 | Yes |
| 75 | Arcn1 | 10446 | -0.302 | -0.1654 | Yes |
| 76 | Snap23 | 10583 | -0.319 | -0.1611 | Yes |
| 77 | Clcn3 | 10628 | -0.323 | -0.1490 | Yes |
| 78 | Ergic3 | 10631 | -0.324 | -0.1335 | Yes |
| 79 | Rab5a | 10642 | -0.325 | -0.1186 | Yes |
| 80 | Napa | 10839 | -0.353 | -0.1175 | Yes |
| 81 | Zw10 | 10921 | -0.363 | -0.1066 | Yes |
| 82 | Scamp3 | 11171 | -0.400 | -0.1075 | Yes |
| 83 | Cope | 11489 | -0.449 | -0.1117 | Yes |
| 84 | Rab14 | 11780 | -0.515 | -0.1104 | Yes |
| 85 | Galc | 11892 | -0.546 | -0.0930 | Yes |
| 86 | Tom1l1 | 11985 | -0.582 | -0.0724 | Yes |
| 87 | Sspn | 12043 | -0.605 | -0.0478 | Yes |
| 88 | Ykt6 | 12143 | -0.662 | -0.0238 | Yes |
| 89 | Atp6v1h | 12297 | -0.862 | 0.0054 | Yes |