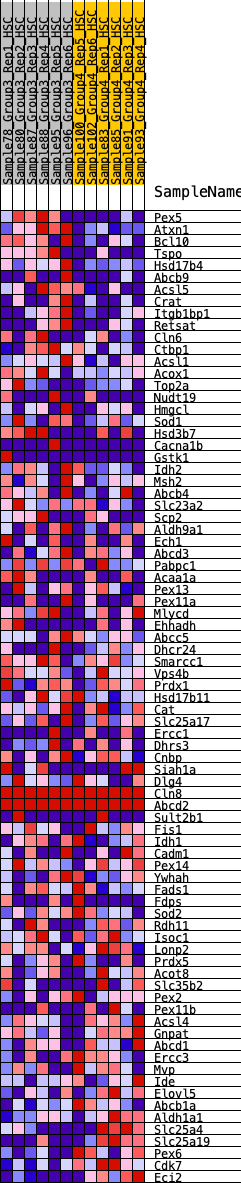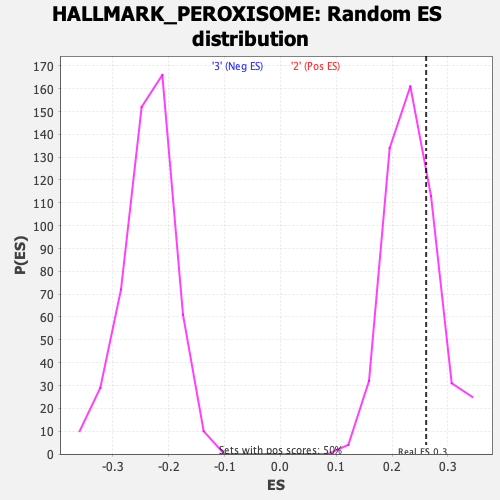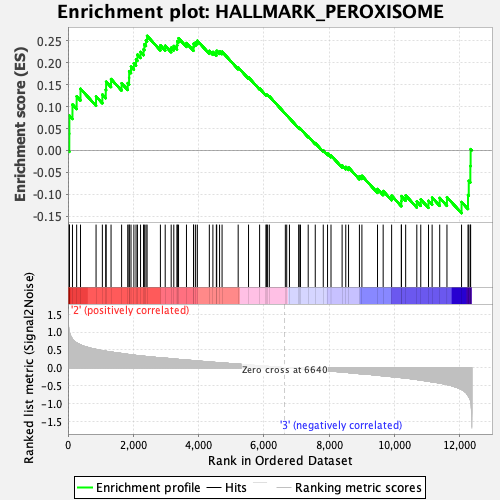
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.26094007 |
| Normalized Enrichment Score (NES) | 1.1030288 |
| Nominal p-value | 0.276 |
| FDR q-value | 0.5636631 |
| FWER p-Value | 0.987 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 40 | 0.994 | 0.0382 | Yes |
| 2 | Atxn1 | 42 | 0.984 | 0.0793 | Yes |
| 3 | Bcl10 | 137 | 0.790 | 0.1046 | Yes |
| 4 | Tspo | 265 | 0.693 | 0.1232 | Yes |
| 5 | Hsd17b4 | 383 | 0.639 | 0.1404 | Yes |
| 6 | Abcb9 | 859 | 0.511 | 0.1231 | Yes |
| 7 | Acsl5 | 1052 | 0.476 | 0.1273 | Yes |
| 8 | Crat | 1156 | 0.460 | 0.1382 | Yes |
| 9 | Itgb1bp1 | 1167 | 0.459 | 0.1565 | Yes |
| 10 | Retsat | 1318 | 0.439 | 0.1626 | Yes |
| 11 | Cln6 | 1641 | 0.397 | 0.1530 | Yes |
| 12 | Ctbp1 | 1829 | 0.373 | 0.1533 | Yes |
| 13 | Acsl1 | 1871 | 0.366 | 0.1653 | Yes |
| 14 | Acox1 | 1873 | 0.366 | 0.1805 | Yes |
| 15 | Top2a | 1929 | 0.359 | 0.1910 | Yes |
| 16 | Nudt19 | 2017 | 0.349 | 0.1985 | Yes |
| 17 | Hmgcl | 2083 | 0.343 | 0.2076 | Yes |
| 18 | Sod1 | 2126 | 0.338 | 0.2183 | Yes |
| 19 | Hsd3b7 | 2221 | 0.328 | 0.2243 | Yes |
| 20 | Cacna1b | 2313 | 0.319 | 0.2302 | Yes |
| 21 | Gstk1 | 2337 | 0.318 | 0.2417 | Yes |
| 22 | Idh2 | 2386 | 0.313 | 0.2508 | Yes |
| 23 | Msh2 | 2421 | 0.308 | 0.2609 | Yes |
| 24 | Abcb4 | 2829 | 0.277 | 0.2394 | No |
| 25 | Slc23a2 | 2976 | 0.264 | 0.2385 | No |
| 26 | Scp2 | 3159 | 0.251 | 0.2342 | No |
| 27 | Aldh9a1 | 3239 | 0.243 | 0.2379 | No |
| 28 | Ech1 | 3337 | 0.234 | 0.2398 | No |
| 29 | Abcd3 | 3347 | 0.234 | 0.2488 | No |
| 30 | Pabpc1 | 3384 | 0.231 | 0.2555 | No |
| 31 | Acaa1a | 3627 | 0.209 | 0.2445 | No |
| 32 | Pex13 | 3843 | 0.196 | 0.2352 | No |
| 33 | Pex11a | 3847 | 0.195 | 0.2431 | No |
| 34 | Mlycd | 3907 | 0.191 | 0.2463 | No |
| 35 | Ehhadh | 3960 | 0.188 | 0.2499 | No |
| 36 | Abcc5 | 4327 | 0.160 | 0.2268 | No |
| 37 | Dhcr24 | 4436 | 0.151 | 0.2243 | No |
| 38 | Smarcc1 | 4542 | 0.144 | 0.2218 | No |
| 39 | Vps4b | 4553 | 0.143 | 0.2269 | No |
| 40 | Prdx1 | 4640 | 0.137 | 0.2256 | No |
| 41 | Hsd17b11 | 4717 | 0.132 | 0.2249 | No |
| 42 | Cat | 5210 | 0.098 | 0.1890 | No |
| 43 | Slc25a17 | 5527 | 0.076 | 0.1665 | No |
| 44 | Ercc1 | 5866 | 0.050 | 0.1410 | No |
| 45 | Dhrs3 | 6060 | 0.040 | 0.1270 | No |
| 46 | Cnbp | 6081 | 0.039 | 0.1270 | No |
| 47 | Siah1a | 6111 | 0.036 | 0.1261 | No |
| 48 | Dlg4 | 6164 | 0.033 | 0.1233 | No |
| 49 | Cln8 | 6654 | 0.000 | 0.0835 | No |
| 50 | Abcd2 | 6696 | 0.000 | 0.0801 | No |
| 51 | Sult2b1 | 6780 | -0.002 | 0.0735 | No |
| 52 | Fis1 | 7064 | -0.022 | 0.0514 | No |
| 53 | Idh1 | 7076 | -0.023 | 0.0515 | No |
| 54 | Cadm1 | 7118 | -0.027 | 0.0492 | No |
| 55 | Pex14 | 7354 | -0.042 | 0.0318 | No |
| 56 | Ywhah | 7570 | -0.058 | 0.0168 | No |
| 57 | Fads1 | 7815 | -0.076 | 0.0001 | No |
| 58 | Fdps | 7946 | -0.086 | -0.0069 | No |
| 59 | Sod2 | 8052 | -0.096 | -0.0115 | No |
| 60 | Rdh11 | 8392 | -0.122 | -0.0340 | No |
| 61 | Isoc1 | 8504 | -0.130 | -0.0376 | No |
| 62 | Lonp2 | 8589 | -0.136 | -0.0387 | No |
| 63 | Prdx5 | 8922 | -0.166 | -0.0588 | No |
| 64 | Acot8 | 8998 | -0.171 | -0.0578 | No |
| 65 | Slc35b2 | 9476 | -0.210 | -0.0878 | No |
| 66 | Pex2 | 9649 | -0.225 | -0.0924 | No |
| 67 | Pex11b | 9908 | -0.251 | -0.1030 | No |
| 68 | Acsl4 | 10205 | -0.278 | -0.1155 | No |
| 69 | Gnpat | 10209 | -0.278 | -0.1041 | No |
| 70 | Abcd1 | 10338 | -0.290 | -0.1024 | No |
| 71 | Ercc3 | 10679 | -0.331 | -0.1163 | No |
| 72 | Mvp | 10800 | -0.348 | -0.1115 | No |
| 73 | Ide | 11037 | -0.381 | -0.1148 | No |
| 74 | Elovl5 | 11148 | -0.394 | -0.1073 | No |
| 75 | Abcb1a | 11380 | -0.429 | -0.1082 | No |
| 76 | Aldh1a1 | 11602 | -0.471 | -0.1065 | No |
| 77 | Slc25a4 | 12049 | -0.607 | -0.1174 | No |
| 78 | Slc25a19 | 12247 | -0.768 | -0.1014 | No |
| 79 | Pex6 | 12270 | -0.814 | -0.0692 | No |
| 80 | Cdk7 | 12318 | -0.901 | -0.0354 | No |
| 81 | Eci2 | 12327 | -0.933 | 0.0029 | No |