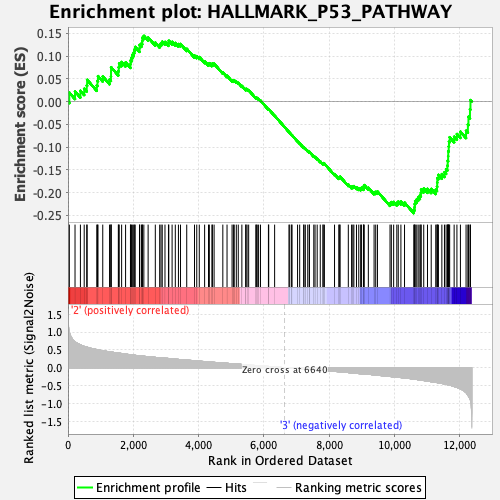
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_P53_PATHWAY |
| Enrichment Score (ES) | -0.24486181 |
| Normalized Enrichment Score (NES) | -1.0485612 |
| Nominal p-value | 0.3838384 |
| FDR q-value | 0.886895 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rchy1 | 41 | 0.993 | 0.0197 | No |
| 2 | Tgfb1 | 215 | 0.719 | 0.0223 | No |
| 3 | Ctsf | 380 | 0.641 | 0.0238 | No |
| 4 | Ctsd | 496 | 0.596 | 0.0282 | No |
| 5 | Abat | 574 | 0.577 | 0.0353 | No |
| 6 | Baiap2 | 586 | 0.573 | 0.0478 | No |
| 7 | Ccnd2 | 881 | 0.508 | 0.0355 | No |
| 8 | Dram1 | 902 | 0.502 | 0.0455 | No |
| 9 | Rrp8 | 918 | 0.499 | 0.0559 | No |
| 10 | Cyfip2 | 1064 | 0.474 | 0.0551 | No |
| 11 | Ccnk | 1272 | 0.444 | 0.0485 | No |
| 12 | Gm2a | 1314 | 0.439 | 0.0553 | No |
| 13 | Retsat | 1318 | 0.439 | 0.0653 | No |
| 14 | Pitpnc1 | 1319 | 0.439 | 0.0755 | No |
| 15 | F2r | 1541 | 0.409 | 0.0669 | No |
| 16 | Dcxr | 1553 | 0.407 | 0.0755 | No |
| 17 | Pcna | 1566 | 0.406 | 0.0839 | No |
| 18 | Rhbdf2 | 1642 | 0.397 | 0.0870 | No |
| 19 | Sesn1 | 1763 | 0.380 | 0.0860 | No |
| 20 | Trp53 | 1912 | 0.360 | 0.0823 | No |
| 21 | Dgka | 1916 | 0.360 | 0.0904 | No |
| 22 | Ddb2 | 1946 | 0.358 | 0.0963 | No |
| 23 | Plxnb2 | 1969 | 0.354 | 0.1028 | No |
| 24 | Tob1 | 2012 | 0.350 | 0.1075 | No |
| 25 | Slc35d1 | 2031 | 0.348 | 0.1141 | No |
| 26 | Casp1 | 2057 | 0.346 | 0.1201 | No |
| 27 | Zfp36l1 | 2191 | 0.332 | 0.1169 | No |
| 28 | Ldhb | 2193 | 0.331 | 0.1246 | No |
| 29 | S100a10 | 2248 | 0.326 | 0.1277 | No |
| 30 | Fbxw7 | 2273 | 0.323 | 0.1333 | No |
| 31 | Mapkapk3 | 2277 | 0.323 | 0.1405 | No |
| 32 | Ier5 | 2322 | 0.319 | 0.1443 | No |
| 33 | Ier3 | 2453 | 0.305 | 0.1408 | No |
| 34 | Ccp110 | 2673 | 0.288 | 0.1296 | No |
| 35 | Hdac3 | 2807 | 0.279 | 0.1252 | No |
| 36 | Csrnp2 | 2847 | 0.276 | 0.1284 | No |
| 37 | Ndrg1 | 2885 | 0.273 | 0.1317 | No |
| 38 | Acvr1b | 2969 | 0.264 | 0.1311 | No |
| 39 | Stom | 3076 | 0.258 | 0.1284 | No |
| 40 | Ptpre | 3086 | 0.257 | 0.1336 | No |
| 41 | H1f2 | 3187 | 0.249 | 0.1312 | No |
| 42 | St14 | 3285 | 0.239 | 0.1288 | No |
| 43 | Ddit4 | 3381 | 0.232 | 0.1264 | No |
| 44 | Zmat3 | 3444 | 0.225 | 0.1266 | No |
| 45 | Klf4 | 3635 | 0.209 | 0.1159 | No |
| 46 | Irak1 | 3873 | 0.194 | 0.1010 | No |
| 47 | Iscu | 3942 | 0.189 | 0.0998 | No |
| 48 | App | 4019 | 0.183 | 0.0978 | No |
| 49 | Kif13b | 4183 | 0.171 | 0.0885 | No |
| 50 | Bax | 4302 | 0.161 | 0.0825 | No |
| 51 | Abcc5 | 4327 | 0.160 | 0.0843 | No |
| 52 | Trib3 | 4407 | 0.153 | 0.0814 | No |
| 53 | Vamp8 | 4421 | 0.152 | 0.0839 | No |
| 54 | Ccng1 | 4473 | 0.149 | 0.0832 | No |
| 55 | Prmt2 | 4740 | 0.130 | 0.0644 | No |
| 56 | Blcap | 4869 | 0.122 | 0.0567 | No |
| 57 | Pidd1 | 5023 | 0.111 | 0.0468 | No |
| 58 | H2aj | 5064 | 0.109 | 0.0461 | No |
| 59 | Klk8 | 5089 | 0.107 | 0.0466 | No |
| 60 | Abhd4 | 5153 | 0.102 | 0.0438 | No |
| 61 | Hint1 | 5213 | 0.098 | 0.0412 | No |
| 62 | Trafd1 | 5323 | 0.090 | 0.0344 | No |
| 63 | Mdm2 | 5441 | 0.081 | 0.0267 | No |
| 64 | Rxra | 5448 | 0.081 | 0.0281 | No |
| 65 | Vwa5a | 5482 | 0.079 | 0.0272 | No |
| 66 | Def6 | 5531 | 0.076 | 0.0250 | No |
| 67 | Ercc5 | 5751 | 0.059 | 0.0085 | No |
| 68 | Fuca1 | 5767 | 0.058 | 0.0086 | No |
| 69 | Zbtb16 | 5793 | 0.056 | 0.0079 | No |
| 70 | Ninj1 | 5835 | 0.052 | 0.0057 | No |
| 71 | Rab40c | 5893 | 0.049 | 0.0022 | No |
| 72 | Ralgds | 6136 | 0.035 | -0.0168 | No |
| 73 | Prkab1 | 6145 | 0.034 | -0.0167 | No |
| 74 | Osgin1 | 6326 | 0.021 | -0.0309 | No |
| 75 | Rps12 | 6765 | -0.001 | -0.0668 | No |
| 76 | Tcn2 | 6789 | -0.003 | -0.0686 | No |
| 77 | Tprkb | 6847 | -0.007 | -0.0731 | No |
| 78 | Mxd4 | 6857 | -0.008 | -0.0736 | No |
| 79 | Pmm1 | 7028 | -0.021 | -0.0871 | No |
| 80 | Rb1 | 7093 | -0.024 | -0.0918 | No |
| 81 | Mknk2 | 7215 | -0.032 | -0.1009 | No |
| 82 | Fam162a | 7225 | -0.033 | -0.1009 | No |
| 83 | Sertad3 | 7266 | -0.037 | -0.1033 | No |
| 84 | Pvt1 | 7340 | -0.041 | -0.1084 | No |
| 85 | Wwp1 | 7389 | -0.046 | -0.1112 | No |
| 86 | Upp1 | 7390 | -0.046 | -0.1102 | No |
| 87 | Nol8 | 7525 | -0.055 | -0.1198 | No |
| 88 | Rad9a | 7563 | -0.058 | -0.1215 | No |
| 89 | Hspa4l | 7629 | -0.062 | -0.1254 | No |
| 90 | Tap1 | 7723 | -0.069 | -0.1314 | No |
| 91 | Fgf13 | 7806 | -0.076 | -0.1364 | No |
| 92 | Tm4sf1 | 7812 | -0.076 | -0.1350 | No |
| 93 | Pom121 | 7850 | -0.079 | -0.1362 | No |
| 94 | Ppp1r15a | 8160 | -0.103 | -0.1591 | No |
| 95 | Socs1 | 8290 | -0.113 | -0.1671 | No |
| 96 | Steap3 | 8318 | -0.115 | -0.1666 | No |
| 97 | Vdr | 8329 | -0.116 | -0.1647 | No |
| 98 | Ei24 | 8582 | -0.136 | -0.1822 | No |
| 99 | Foxo3 | 8682 | -0.145 | -0.1869 | No |
| 100 | Cgrrf1 | 8720 | -0.148 | -0.1865 | No |
| 101 | Sec61a1 | 8752 | -0.151 | -0.1855 | No |
| 102 | Txnip | 8832 | -0.160 | -0.1883 | No |
| 103 | Elp1 | 8902 | -0.164 | -0.1901 | No |
| 104 | Apaf1 | 8960 | -0.169 | -0.1909 | No |
| 105 | Bak1 | 8988 | -0.171 | -0.1891 | No |
| 106 | Aen | 9047 | -0.174 | -0.1898 | No |
| 107 | Cdkn2aip | 9051 | -0.174 | -0.1860 | No |
| 108 | Ephx1 | 9069 | -0.176 | -0.1833 | No |
| 109 | Btg1 | 9195 | -0.188 | -0.1892 | No |
| 110 | Tspyl2 | 9371 | -0.200 | -0.1989 | No |
| 111 | Slc3a2 | 9421 | -0.205 | -0.1981 | No |
| 112 | Hexim1 | 9474 | -0.210 | -0.1975 | No |
| 113 | Mxd1 | 9859 | -0.246 | -0.2232 | No |
| 114 | Ptpn14 | 9899 | -0.250 | -0.2206 | No |
| 115 | Wrap73 | 9970 | -0.258 | -0.2203 | No |
| 116 | Jag2 | 10067 | -0.265 | -0.2220 | No |
| 117 | Cd81 | 10111 | -0.268 | -0.2193 | No |
| 118 | Polh | 10192 | -0.276 | -0.2194 | No |
| 119 | Sp1 | 10303 | -0.288 | -0.2218 | No |
| 120 | Tpd52l1 | 10586 | -0.319 | -0.2374 | Yes |
| 121 | Eps8l2 | 10611 | -0.322 | -0.2319 | Yes |
| 122 | Fos | 10616 | -0.322 | -0.2248 | Yes |
| 123 | Tsc22d1 | 10629 | -0.324 | -0.2182 | Yes |
| 124 | Jun | 10678 | -0.331 | -0.2144 | Yes |
| 125 | Notch1 | 10721 | -0.337 | -0.2101 | Yes |
| 126 | Cd82 | 10773 | -0.344 | -0.2062 | Yes |
| 127 | Plk2 | 10803 | -0.349 | -0.2005 | Yes |
| 128 | Ppm1d | 10811 | -0.350 | -0.1929 | Yes |
| 129 | Rnf19b | 10891 | -0.360 | -0.1910 | Yes |
| 130 | Ccnd3 | 11006 | -0.377 | -0.1916 | Yes |
| 131 | Hbegf | 11120 | -0.393 | -0.1917 | Yes |
| 132 | Rack1 | 11259 | -0.411 | -0.1934 | Yes |
| 133 | Sat1 | 11297 | -0.417 | -0.1868 | Yes |
| 134 | Tm7sf3 | 11304 | -0.418 | -0.1775 | Yes |
| 135 | Fdxr | 11308 | -0.419 | -0.1680 | Yes |
| 136 | Ip6k2 | 11341 | -0.423 | -0.1608 | Yes |
| 137 | Tgfa | 11446 | -0.440 | -0.1591 | Yes |
| 138 | Rap2b | 11526 | -0.456 | -0.1550 | Yes |
| 139 | Slc19a2 | 11576 | -0.465 | -0.1482 | Yes |
| 140 | Ddit3 | 11617 | -0.474 | -0.1404 | Yes |
| 141 | Ak1 | 11625 | -0.477 | -0.1299 | Yes |
| 142 | H2ac25 | 11639 | -0.479 | -0.1198 | Yes |
| 143 | Fas | 11642 | -0.481 | -0.1088 | Yes |
| 144 | Rpl18 | 11656 | -0.483 | -0.0986 | Yes |
| 145 | Ada | 11664 | -0.485 | -0.0879 | Yes |
| 146 | Irag2 | 11685 | -0.490 | -0.0782 | Yes |
| 147 | Procr | 11822 | -0.529 | -0.0770 | Yes |
| 148 | Dnttip2 | 11907 | -0.549 | -0.0711 | Yes |
| 149 | Rps27l | 12014 | -0.592 | -0.0660 | Yes |
| 150 | Ankra2 | 12187 | -0.699 | -0.0638 | Yes |
| 151 | Coq8a | 12243 | -0.764 | -0.0506 | Yes |
| 152 | Xpc | 12260 | -0.800 | -0.0333 | Yes |
| 153 | Nupr1 | 12307 | -0.879 | -0.0166 | Yes |
| 154 | Rpl36 | 12322 | -0.907 | 0.0034 | Yes |

