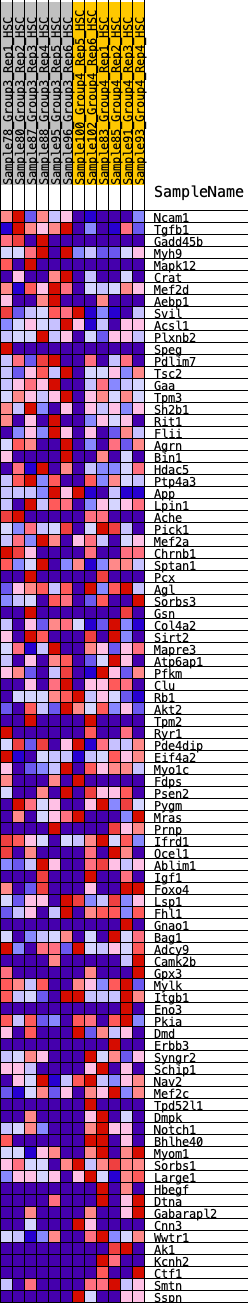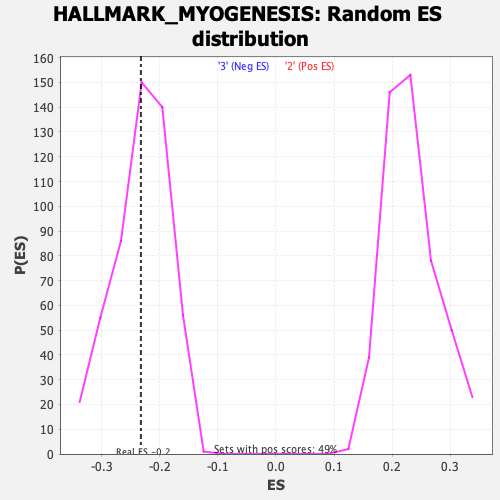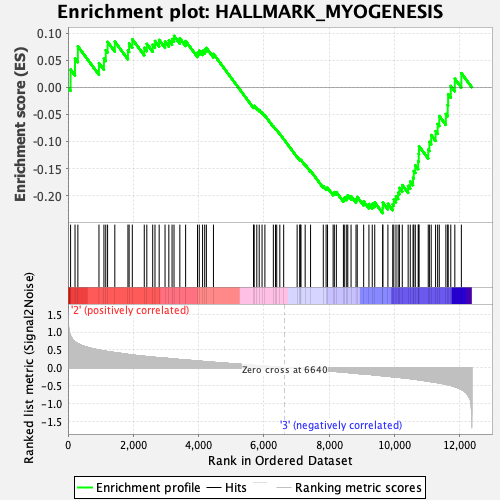
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.23232408 |
| Normalized Enrichment Score (NES) | -1.0062095 |
| Nominal p-value | 0.42632613 |
| FDR q-value | 0.8511964 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ncam1 | 79 | 0.894 | 0.0327 | No |
| 2 | Tgfb1 | 215 | 0.719 | 0.0532 | No |
| 3 | Gadd45b | 302 | 0.672 | 0.0756 | No |
| 4 | Myh9 | 948 | 0.494 | 0.0447 | No |
| 5 | Mapk12 | 1100 | 0.468 | 0.0529 | No |
| 6 | Crat | 1156 | 0.460 | 0.0685 | No |
| 7 | Mef2d | 1212 | 0.452 | 0.0838 | No |
| 8 | Aebp1 | 1433 | 0.420 | 0.0843 | No |
| 9 | Svil | 1835 | 0.372 | 0.0679 | No |
| 10 | Acsl1 | 1871 | 0.366 | 0.0811 | No |
| 11 | Plxnb2 | 1969 | 0.354 | 0.0887 | No |
| 12 | Speg | 2335 | 0.318 | 0.0729 | No |
| 13 | Pdlim7 | 2413 | 0.309 | 0.0801 | No |
| 14 | Tsc2 | 2593 | 0.294 | 0.0784 | No |
| 15 | Gaa | 2663 | 0.289 | 0.0854 | No |
| 16 | Tpm3 | 2791 | 0.280 | 0.0874 | No |
| 17 | Sh2b1 | 2971 | 0.264 | 0.0843 | No |
| 18 | Rit1 | 3087 | 0.257 | 0.0862 | No |
| 19 | Flii | 3186 | 0.249 | 0.0891 | No |
| 20 | Agrn | 3246 | 0.242 | 0.0949 | No |
| 21 | Bin1 | 3424 | 0.227 | 0.0904 | No |
| 22 | Hdac5 | 3601 | 0.212 | 0.0853 | No |
| 23 | Ptp4a3 | 3965 | 0.187 | 0.0640 | No |
| 24 | App | 4019 | 0.183 | 0.0677 | No |
| 25 | Lpin1 | 4120 | 0.175 | 0.0672 | No |
| 26 | Ache | 4185 | 0.171 | 0.0694 | No |
| 27 | Pick1 | 4236 | 0.166 | 0.0726 | No |
| 28 | Mef2a | 4453 | 0.150 | 0.0616 | No |
| 29 | Chrnb1 | 5679 | 0.064 | -0.0354 | No |
| 30 | Sptan1 | 5700 | 0.063 | -0.0343 | No |
| 31 | Pcx | 5780 | 0.057 | -0.0382 | No |
| 32 | Agl | 5854 | 0.051 | -0.0419 | No |
| 33 | Sorbs3 | 5941 | 0.046 | -0.0469 | No |
| 34 | Gsn | 6028 | 0.041 | -0.0522 | No |
| 35 | Col4a2 | 6287 | 0.023 | -0.0722 | No |
| 36 | Sirt2 | 6352 | 0.019 | -0.0766 | No |
| 37 | Mapre3 | 6390 | 0.016 | -0.0789 | No |
| 38 | Atp6ap1 | 6485 | 0.010 | -0.0861 | No |
| 39 | Pfkm | 6603 | 0.002 | -0.0955 | No |
| 40 | Clu | 7015 | -0.020 | -0.1282 | No |
| 41 | Rb1 | 7093 | -0.024 | -0.1334 | No |
| 42 | Akt2 | 7114 | -0.026 | -0.1339 | No |
| 43 | Tpm2 | 7133 | -0.027 | -0.1341 | No |
| 44 | Ryr1 | 7261 | -0.037 | -0.1429 | No |
| 45 | Pde4dip | 7426 | -0.048 | -0.1541 | No |
| 46 | Eif4a2 | 7816 | -0.076 | -0.1825 | No |
| 47 | Myo1c | 7902 | -0.083 | -0.1858 | No |
| 48 | Fdps | 7946 | -0.086 | -0.1855 | No |
| 49 | Psen2 | 8117 | -0.099 | -0.1950 | No |
| 50 | Pygm | 8154 | -0.103 | -0.1935 | No |
| 51 | Mras | 8216 | -0.107 | -0.1937 | No |
| 52 | Prnp | 8432 | -0.125 | -0.2058 | No |
| 53 | Ifrd1 | 8472 | -0.128 | -0.2034 | No |
| 54 | Ocel1 | 8534 | -0.132 | -0.2026 | No |
| 55 | Ablim1 | 8566 | -0.134 | -0.1992 | No |
| 56 | Igf1 | 8666 | -0.144 | -0.2010 | No |
| 57 | Foxo4 | 8818 | -0.158 | -0.2064 | No |
| 58 | Lsp1 | 8855 | -0.160 | -0.2023 | No |
| 59 | Fhl1 | 9052 | -0.174 | -0.2106 | No |
| 60 | Gnao1 | 9213 | -0.188 | -0.2154 | No |
| 61 | Bag1 | 9315 | -0.195 | -0.2151 | No |
| 62 | Adcy9 | 9391 | -0.201 | -0.2124 | No |
| 63 | Camk2b | 9636 | -0.224 | -0.2225 | Yes |
| 64 | Gpx3 | 9638 | -0.224 | -0.2128 | Yes |
| 65 | Mylk | 9794 | -0.238 | -0.2150 | Yes |
| 66 | Itgb1 | 9942 | -0.255 | -0.2158 | Yes |
| 67 | Eno3 | 9975 | -0.258 | -0.2072 | Yes |
| 68 | Pkia | 10042 | -0.262 | -0.2011 | Yes |
| 69 | Dmd | 10108 | -0.268 | -0.1946 | Yes |
| 70 | Erbb3 | 10147 | -0.274 | -0.1858 | Yes |
| 71 | Syngr2 | 10234 | -0.281 | -0.1805 | Yes |
| 72 | Schip1 | 10414 | -0.298 | -0.1820 | Yes |
| 73 | Nav2 | 10479 | -0.306 | -0.1738 | Yes |
| 74 | Mef2c | 10564 | -0.317 | -0.1668 | Yes |
| 75 | Tpd52l1 | 10586 | -0.319 | -0.1546 | Yes |
| 76 | Dmpk | 10633 | -0.324 | -0.1441 | Yes |
| 77 | Notch1 | 10721 | -0.337 | -0.1365 | Yes |
| 78 | Bhlhe40 | 10738 | -0.339 | -0.1230 | Yes |
| 79 | Myom1 | 10746 | -0.341 | -0.1086 | Yes |
| 80 | Sorbs1 | 11029 | -0.380 | -0.1150 | Yes |
| 81 | Large1 | 11064 | -0.386 | -0.1008 | Yes |
| 82 | Hbegf | 11120 | -0.393 | -0.0881 | Yes |
| 83 | Dtna | 11254 | -0.410 | -0.0810 | Yes |
| 84 | Gabarapl2 | 11317 | -0.419 | -0.0677 | Yes |
| 85 | Cnn3 | 11367 | -0.427 | -0.0530 | Yes |
| 86 | Wwtr1 | 11569 | -0.463 | -0.0492 | Yes |
| 87 | Ak1 | 11625 | -0.477 | -0.0328 | Yes |
| 88 | Kcnh2 | 11638 | -0.479 | -0.0128 | Yes |
| 89 | Ctf1 | 11719 | -0.497 | 0.0025 | Yes |
| 90 | Smtn | 11843 | -0.534 | 0.0158 | Yes |
| 91 | Sspn | 12043 | -0.605 | 0.0261 | Yes |