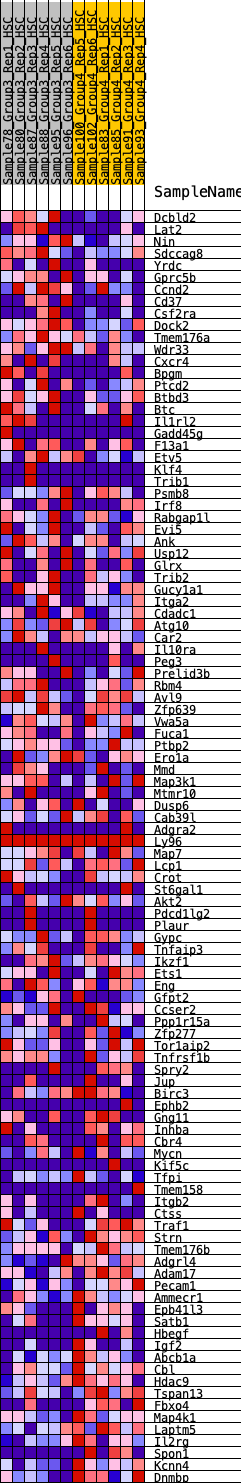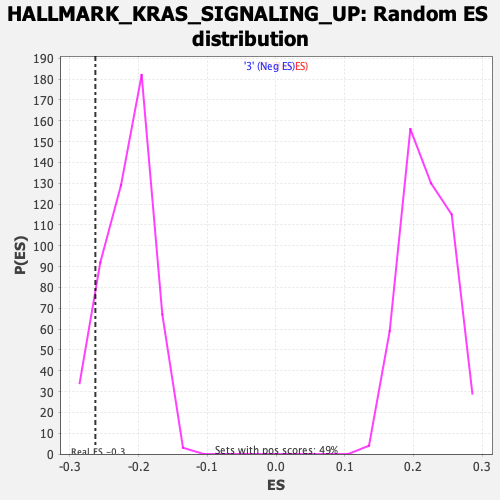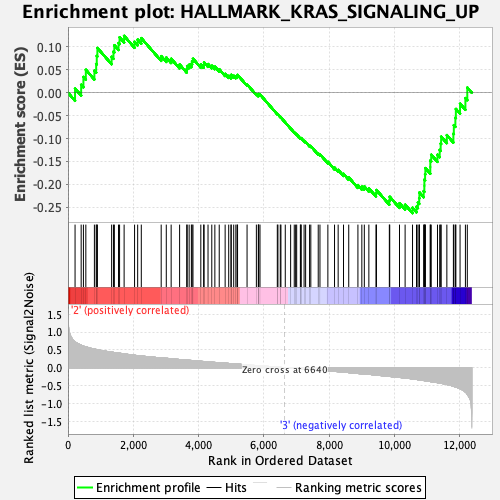
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.26282203 |
| Normalized Enrichment Score (NES) | -1.219251 |
| Nominal p-value | 0.102564104 |
| FDR q-value | 0.9457577 |
| FWER p-Value | 0.914 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcbld2 | 216 | 0.718 | 0.0091 | No |
| 2 | Lat2 | 403 | 0.630 | 0.0175 | No |
| 3 | Nin | 472 | 0.604 | 0.0344 | No |
| 4 | Sdccag8 | 546 | 0.581 | 0.0501 | No |
| 5 | Yrdc | 808 | 0.520 | 0.0482 | No |
| 6 | Gprc5b | 863 | 0.511 | 0.0629 | No |
| 7 | Ccnd2 | 881 | 0.508 | 0.0804 | No |
| 8 | Cd37 | 900 | 0.503 | 0.0977 | No |
| 9 | Csf2ra | 1336 | 0.436 | 0.0784 | No |
| 10 | Dock2 | 1392 | 0.428 | 0.0899 | No |
| 11 | Tmem176a | 1419 | 0.423 | 0.1036 | No |
| 12 | Wdr33 | 1547 | 0.408 | 0.1084 | No |
| 13 | Cxcr4 | 1579 | 0.404 | 0.1209 | No |
| 14 | Bpgm | 1718 | 0.387 | 0.1241 | No |
| 15 | Ptcd2 | 2038 | 0.348 | 0.1110 | No |
| 16 | Btbd3 | 2133 | 0.337 | 0.1159 | No |
| 17 | Btc | 2244 | 0.326 | 0.1191 | No |
| 18 | Il1rl2 | 2853 | 0.276 | 0.0798 | No |
| 19 | Gadd45g | 3009 | 0.261 | 0.0769 | No |
| 20 | F13a1 | 3158 | 0.251 | 0.0741 | No |
| 21 | Etv5 | 3417 | 0.227 | 0.0616 | No |
| 22 | Klf4 | 3635 | 0.209 | 0.0516 | No |
| 23 | Trib1 | 3649 | 0.209 | 0.0584 | No |
| 24 | Psmb8 | 3709 | 0.206 | 0.0612 | No |
| 25 | Irf8 | 3777 | 0.202 | 0.0633 | No |
| 26 | Rabgap1l | 3799 | 0.200 | 0.0690 | No |
| 27 | Evi5 | 3826 | 0.197 | 0.0742 | No |
| 28 | Ank | 4066 | 0.179 | 0.0614 | No |
| 29 | Usp12 | 4153 | 0.173 | 0.0609 | No |
| 30 | Glrx | 4166 | 0.172 | 0.0663 | No |
| 31 | Trib2 | 4287 | 0.162 | 0.0625 | No |
| 32 | Gucy1a1 | 4402 | 0.153 | 0.0590 | No |
| 33 | Itga2 | 4496 | 0.147 | 0.0569 | No |
| 34 | Cdadc1 | 4630 | 0.138 | 0.0512 | No |
| 35 | Atg10 | 4811 | 0.126 | 0.0411 | No |
| 36 | Car2 | 4920 | 0.118 | 0.0368 | No |
| 37 | Il10ra | 4991 | 0.113 | 0.0353 | No |
| 38 | Peg3 | 4996 | 0.113 | 0.0391 | No |
| 39 | Prelid3b | 5073 | 0.108 | 0.0370 | No |
| 40 | Rbm4 | 5139 | 0.103 | 0.0355 | No |
| 41 | Avl9 | 5178 | 0.100 | 0.0361 | No |
| 42 | Zfp639 | 5191 | 0.099 | 0.0389 | No |
| 43 | Vwa5a | 5482 | 0.079 | 0.0182 | No |
| 44 | Fuca1 | 5767 | 0.058 | -0.0029 | No |
| 45 | Ptbp2 | 5828 | 0.053 | -0.0058 | No |
| 46 | Ero1a | 5830 | 0.053 | -0.0039 | No |
| 47 | Mmd | 5839 | 0.052 | -0.0026 | No |
| 48 | Map3k1 | 5878 | 0.050 | -0.0039 | No |
| 49 | Mtmr10 | 6408 | 0.015 | -0.0465 | No |
| 50 | Dusp6 | 6427 | 0.014 | -0.0474 | No |
| 51 | Cab39l | 6499 | 0.008 | -0.0529 | No |
| 52 | Adgra2 | 6519 | 0.007 | -0.0542 | No |
| 53 | Ly96 | 6652 | 0.000 | -0.0650 | No |
| 54 | Map7 | 6816 | -0.005 | -0.0781 | No |
| 55 | Lcp1 | 6927 | -0.013 | -0.0866 | No |
| 56 | Crot | 6969 | -0.016 | -0.0893 | No |
| 57 | St6gal1 | 7001 | -0.019 | -0.0911 | No |
| 58 | Akt2 | 7114 | -0.026 | -0.0993 | No |
| 59 | Pdcd1lg2 | 7128 | -0.027 | -0.0993 | No |
| 60 | Plaur | 7138 | -0.027 | -0.0991 | No |
| 61 | Gypc | 7228 | -0.033 | -0.1051 | No |
| 62 | Tnfaip3 | 7275 | -0.038 | -0.1074 | No |
| 63 | Ikzf1 | 7396 | -0.046 | -0.1155 | No |
| 64 | Ets1 | 7434 | -0.049 | -0.1167 | No |
| 65 | Eng | 7661 | -0.064 | -0.1327 | No |
| 66 | Gfpt2 | 7714 | -0.068 | -0.1344 | No |
| 67 | Ccser2 | 7956 | -0.087 | -0.1508 | No |
| 68 | Ppp1r15a | 8160 | -0.103 | -0.1636 | No |
| 69 | Zfp277 | 8271 | -0.111 | -0.1684 | No |
| 70 | Tor1aip2 | 8435 | -0.125 | -0.1770 | No |
| 71 | Tnfrsf1b | 8595 | -0.137 | -0.1849 | No |
| 72 | Spry2 | 8876 | -0.162 | -0.2017 | No |
| 73 | Jup | 8996 | -0.171 | -0.2050 | No |
| 74 | Birc3 | 9072 | -0.176 | -0.2046 | No |
| 75 | Ephb2 | 9211 | -0.188 | -0.2088 | No |
| 76 | Gng11 | 9429 | -0.205 | -0.2189 | No |
| 77 | Inhba | 9444 | -0.206 | -0.2123 | No |
| 78 | Cbr4 | 9838 | -0.243 | -0.2353 | No |
| 79 | Mycn | 9850 | -0.244 | -0.2271 | No |
| 80 | Kif5c | 10149 | -0.274 | -0.2412 | No |
| 81 | Tfpi | 10321 | -0.289 | -0.2444 | No |
| 82 | Tmem158 | 10548 | -0.316 | -0.2511 | Yes |
| 83 | Itgb2 | 10670 | -0.329 | -0.2487 | Yes |
| 84 | Ctss | 10713 | -0.336 | -0.2396 | Yes |
| 85 | Traf1 | 10753 | -0.342 | -0.2300 | Yes |
| 86 | Strn | 10759 | -0.342 | -0.2177 | Yes |
| 87 | Tmem176b | 10889 | -0.360 | -0.2148 | Yes |
| 88 | Adgrl4 | 10910 | -0.361 | -0.2030 | Yes |
| 89 | Adam17 | 10911 | -0.361 | -0.1895 | Yes |
| 90 | Pecam1 | 10933 | -0.365 | -0.1776 | Yes |
| 91 | Ammecr1 | 10941 | -0.367 | -0.1645 | Yes |
| 92 | Epb41l3 | 11092 | -0.388 | -0.1623 | Yes |
| 93 | Satb1 | 11093 | -0.388 | -0.1478 | Yes |
| 94 | Hbegf | 11120 | -0.393 | -0.1353 | Yes |
| 95 | Igf2 | 11312 | -0.419 | -0.1353 | Yes |
| 96 | Abcb1a | 11380 | -0.429 | -0.1248 | Yes |
| 97 | Cbl | 11410 | -0.433 | -0.1110 | Yes |
| 98 | Hdac9 | 11424 | -0.435 | -0.0958 | Yes |
| 99 | Tspan13 | 11599 | -0.470 | -0.0925 | Yes |
| 100 | Fbxo4 | 11799 | -0.520 | -0.0894 | Yes |
| 101 | Map4k1 | 11813 | -0.526 | -0.0708 | Yes |
| 102 | Laptm5 | 11860 | -0.538 | -0.0545 | Yes |
| 103 | Il2rg | 11875 | -0.542 | -0.0355 | Yes |
| 104 | Spon1 | 12003 | -0.587 | -0.0239 | Yes |
| 105 | Kcnn4 | 12167 | -0.683 | -0.0117 | Yes |
| 106 | Dnmbp | 12225 | -0.742 | 0.0113 | Yes |