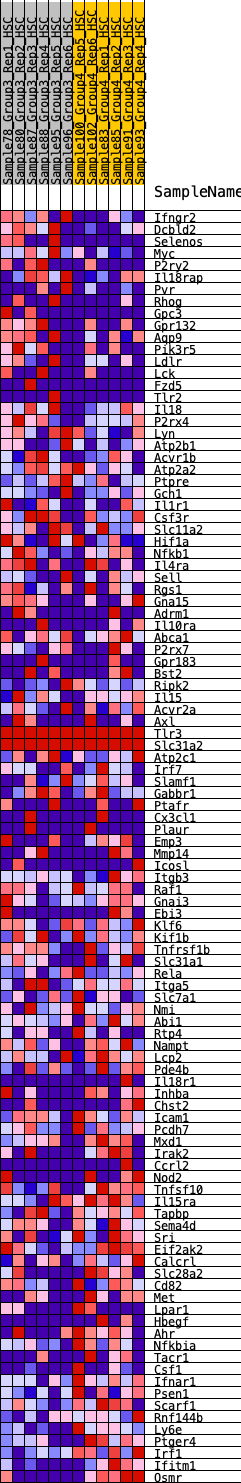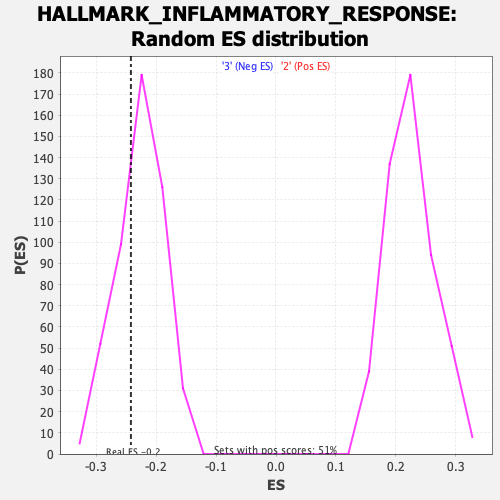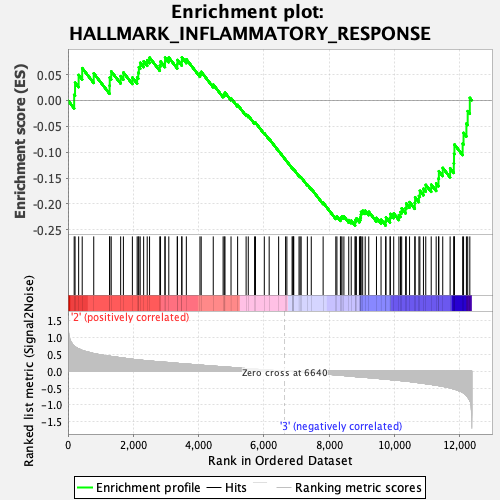
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.24182595 |
| Normalized Enrichment Score (NES) | -1.0725951 |
| Nominal p-value | 0.3089431 |
| FDR q-value | 0.9013487 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ifngr2 | 189 | 0.740 | 0.0110 | No |
| 2 | Dcbld2 | 216 | 0.718 | 0.0344 | No |
| 3 | Selenos | 326 | 0.664 | 0.0492 | No |
| 4 | Myc | 435 | 0.615 | 0.0623 | No |
| 5 | P2ry2 | 788 | 0.523 | 0.0522 | No |
| 6 | Il18rap | 1273 | 0.444 | 0.0286 | No |
| 7 | Pvr | 1278 | 0.443 | 0.0440 | No |
| 8 | Rhog | 1323 | 0.438 | 0.0561 | No |
| 9 | Gpc3 | 1612 | 0.401 | 0.0469 | No |
| 10 | Gpr132 | 1696 | 0.389 | 0.0539 | No |
| 11 | Aqp9 | 1971 | 0.354 | 0.0442 | No |
| 12 | Pik3r5 | 2117 | 0.339 | 0.0444 | No |
| 13 | Ldlr | 2151 | 0.336 | 0.0537 | No |
| 14 | Lck | 2170 | 0.334 | 0.0641 | No |
| 15 | Fzd5 | 2210 | 0.329 | 0.0727 | No |
| 16 | Tlr2 | 2316 | 0.319 | 0.0755 | No |
| 17 | Il18 | 2425 | 0.308 | 0.0777 | No |
| 18 | P2rx4 | 2497 | 0.303 | 0.0827 | No |
| 19 | Lyn | 2811 | 0.278 | 0.0670 | No |
| 20 | Atp2b1 | 2833 | 0.277 | 0.0752 | No |
| 21 | Acvr1b | 2969 | 0.264 | 0.0736 | No |
| 22 | Atp2a2 | 2972 | 0.264 | 0.0828 | No |
| 23 | Ptpre | 3086 | 0.257 | 0.0828 | No |
| 24 | Gch1 | 3346 | 0.234 | 0.0699 | No |
| 25 | Il1r1 | 3349 | 0.233 | 0.0781 | No |
| 26 | Csf3r | 3484 | 0.223 | 0.0751 | No |
| 27 | Slc11a2 | 3486 | 0.223 | 0.0830 | No |
| 28 | Hif1a | 3623 | 0.210 | 0.0794 | No |
| 29 | Nfkb1 | 4040 | 0.182 | 0.0519 | No |
| 30 | Il4ra | 4078 | 0.178 | 0.0552 | No |
| 31 | Sell | 4447 | 0.151 | 0.0306 | No |
| 32 | Rgs1 | 4749 | 0.129 | 0.0107 | No |
| 33 | Gna15 | 4784 | 0.127 | 0.0124 | No |
| 34 | Adrm1 | 4806 | 0.126 | 0.0152 | No |
| 35 | Il10ra | 4991 | 0.113 | 0.0042 | No |
| 36 | Abca1 | 5193 | 0.099 | -0.0087 | No |
| 37 | P2rx7 | 5453 | 0.080 | -0.0269 | No |
| 38 | Gpr183 | 5519 | 0.076 | -0.0295 | No |
| 39 | Bst2 | 5712 | 0.062 | -0.0430 | No |
| 40 | Ripk2 | 5740 | 0.060 | -0.0430 | No |
| 41 | Il15 | 6010 | 0.042 | -0.0635 | No |
| 42 | Acvr2a | 6159 | 0.034 | -0.0744 | No |
| 43 | Axl | 6449 | 0.012 | -0.0975 | No |
| 44 | Tlr3 | 6661 | 0.000 | -0.1147 | No |
| 45 | Slc31a2 | 6701 | 0.000 | -0.1179 | No |
| 46 | Atp2c1 | 6861 | -0.008 | -0.1306 | No |
| 47 | Irf7 | 6882 | -0.010 | -0.1319 | No |
| 48 | Slamf1 | 6907 | -0.012 | -0.1334 | No |
| 49 | Gabbr1 | 6909 | -0.012 | -0.1331 | No |
| 50 | Ptafr | 7073 | -0.023 | -0.1455 | No |
| 51 | Cx3cl1 | 7106 | -0.025 | -0.1473 | No |
| 52 | Plaur | 7138 | -0.027 | -0.1488 | No |
| 53 | Emp3 | 7328 | -0.040 | -0.1628 | No |
| 54 | Mmp14 | 7448 | -0.050 | -0.1707 | No |
| 55 | Icosl | 7811 | -0.076 | -0.1975 | No |
| 56 | Itgb3 | 8199 | -0.106 | -0.2253 | No |
| 57 | Raf1 | 8233 | -0.108 | -0.2242 | No |
| 58 | Gnai3 | 8340 | -0.117 | -0.2286 | No |
| 59 | Ebi3 | 8359 | -0.119 | -0.2259 | No |
| 60 | Klf6 | 8389 | -0.122 | -0.2239 | No |
| 61 | Kif1b | 8447 | -0.126 | -0.2240 | No |
| 62 | Tnfrsf1b | 8595 | -0.137 | -0.2311 | No |
| 63 | Slc31a1 | 8671 | -0.144 | -0.2321 | No |
| 64 | Rela | 8791 | -0.157 | -0.2362 | Yes |
| 65 | Itga5 | 8794 | -0.157 | -0.2308 | Yes |
| 66 | Slc7a1 | 8829 | -0.159 | -0.2279 | Yes |
| 67 | Nmi | 8923 | -0.166 | -0.2296 | Yes |
| 68 | Abi1 | 8951 | -0.168 | -0.2258 | Yes |
| 69 | Rtp4 | 8966 | -0.169 | -0.2209 | Yes |
| 70 | Nampt | 8971 | -0.169 | -0.2152 | Yes |
| 71 | Lcp2 | 9022 | -0.173 | -0.2131 | Yes |
| 72 | Pde4b | 9099 | -0.179 | -0.2129 | Yes |
| 73 | Il18r1 | 9208 | -0.188 | -0.2150 | Yes |
| 74 | Inhba | 9444 | -0.206 | -0.2269 | Yes |
| 75 | Chst2 | 9584 | -0.221 | -0.2303 | Yes |
| 76 | Icam1 | 9725 | -0.232 | -0.2335 | Yes |
| 77 | Pcdh7 | 9735 | -0.232 | -0.2259 | Yes |
| 78 | Mxd1 | 9859 | -0.246 | -0.2272 | Yes |
| 79 | Irak2 | 9870 | -0.247 | -0.2192 | Yes |
| 80 | Ccrl2 | 9972 | -0.258 | -0.2183 | Yes |
| 81 | Nod2 | 10131 | -0.271 | -0.2215 | Yes |
| 82 | Tnfsf10 | 10179 | -0.275 | -0.2156 | Yes |
| 83 | Il15ra | 10216 | -0.279 | -0.2086 | Yes |
| 84 | Tapbp | 10337 | -0.290 | -0.2080 | Yes |
| 85 | Sema4d | 10356 | -0.293 | -0.1990 | Yes |
| 86 | Sri | 10457 | -0.303 | -0.1964 | Yes |
| 87 | Eif2ak2 | 10618 | -0.322 | -0.1980 | Yes |
| 88 | Calcrl | 10627 | -0.322 | -0.1871 | Yes |
| 89 | Slc28a2 | 10744 | -0.340 | -0.1845 | Yes |
| 90 | Cd82 | 10773 | -0.344 | -0.1745 | Yes |
| 91 | Met | 10882 | -0.359 | -0.1705 | Yes |
| 92 | Lpar1 | 10953 | -0.369 | -0.1631 | Yes |
| 93 | Hbegf | 11120 | -0.393 | -0.1627 | Yes |
| 94 | Ahr | 11272 | -0.413 | -0.1603 | Yes |
| 95 | Nfkbia | 11343 | -0.424 | -0.1509 | Yes |
| 96 | Tacr1 | 11355 | -0.424 | -0.1367 | Yes |
| 97 | Csf1 | 11473 | -0.446 | -0.1303 | Yes |
| 98 | Ifnar1 | 11699 | -0.493 | -0.1311 | Yes |
| 99 | Psen1 | 11812 | -0.526 | -0.1215 | Yes |
| 100 | Scarf1 | 11819 | -0.528 | -0.1032 | Yes |
| 101 | Rnf144b | 11831 | -0.530 | -0.0852 | Yes |
| 102 | Ly6e | 12083 | -0.625 | -0.0834 | Yes |
| 103 | Ptger4 | 12110 | -0.640 | -0.0627 | Yes |
| 104 | Irf1 | 12198 | -0.709 | -0.0446 | Yes |
| 105 | Ifitm1 | 12237 | -0.757 | -0.0207 | Yes |
| 106 | Osmr | 12302 | -0.867 | 0.0050 | Yes |