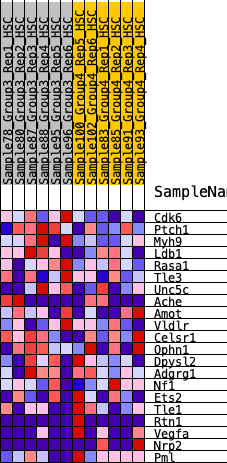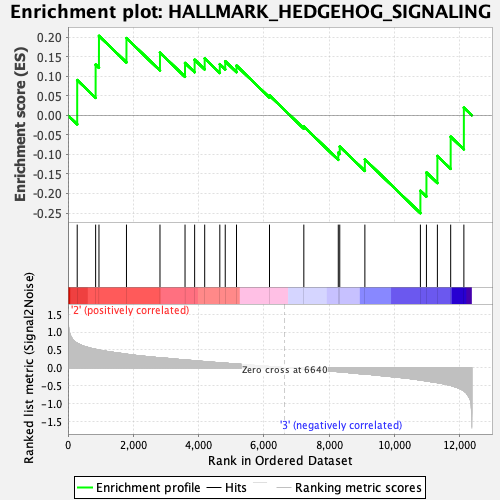
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | -0.2503461 |
| Normalized Enrichment Score (NES) | -0.8015652 |
| Nominal p-value | 0.79476863 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdk6 | 282 | 0.684 | 0.0905 | Yes |
| 2 | Ptch1 | 846 | 0.514 | 0.1300 | Yes |
| 3 | Myh9 | 948 | 0.494 | 0.2036 | Yes |
| 4 | Ldb1 | 1790 | 0.377 | 0.1979 | No |
| 5 | Rasa1 | 2816 | 0.278 | 0.1609 | No |
| 6 | Tle3 | 3583 | 0.214 | 0.1342 | No |
| 7 | Unc5c | 3877 | 0.193 | 0.1425 | No |
| 8 | Ache | 4185 | 0.171 | 0.1460 | No |
| 9 | Amot | 4648 | 0.137 | 0.1311 | No |
| 10 | Vldlr | 4814 | 0.125 | 0.1385 | No |
| 11 | Celsr1 | 5159 | 0.102 | 0.1275 | No |
| 12 | Ophn1 | 6169 | 0.033 | 0.0511 | No |
| 13 | Dpysl2 | 7220 | -0.033 | -0.0285 | No |
| 14 | Adgrg1 | 8275 | -0.111 | -0.0955 | No |
| 15 | Nf1 | 8320 | -0.115 | -0.0799 | No |
| 16 | Ets2 | 9089 | -0.178 | -0.1127 | No |
| 17 | Tle1 | 10789 | -0.347 | -0.1930 | No |
| 18 | Rtn1 | 10974 | -0.372 | -0.1463 | No |
| 19 | Vegfa | 11309 | -0.419 | -0.1040 | No |
| 20 | Nrp2 | 11716 | -0.497 | -0.0545 | No |
| 21 | Pml | 12120 | -0.645 | 0.0197 | No |