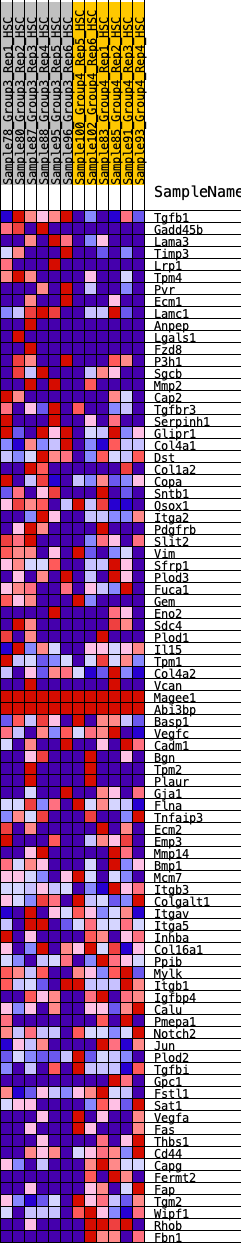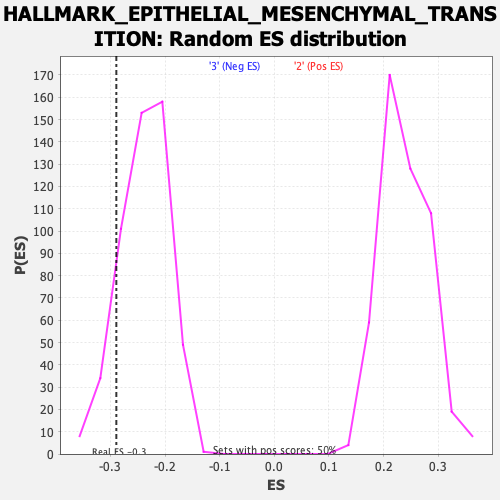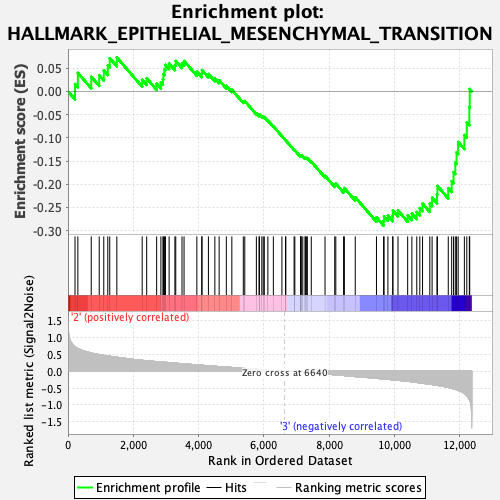
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.28911728 |
| Normalized Enrichment Score (NES) | -1.2126813 |
| Nominal p-value | 0.13690476 |
| FDR q-value | 0.73763174 |
| FWER p-Value | 0.921 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tgfb1 | 215 | 0.719 | 0.0157 | No |
| 2 | Gadd45b | 302 | 0.672 | 0.0398 | No |
| 3 | Lama3 | 711 | 0.540 | 0.0316 | No |
| 4 | Timp3 | 956 | 0.493 | 0.0345 | No |
| 5 | Lrp1 | 1095 | 0.469 | 0.0449 | No |
| 6 | Tpm4 | 1218 | 0.451 | 0.0559 | No |
| 7 | Pvr | 1278 | 0.443 | 0.0715 | No |
| 8 | Ecm1 | 1495 | 0.415 | 0.0731 | No |
| 9 | Lamc1 | 2271 | 0.324 | 0.0250 | No |
| 10 | Anpep | 2409 | 0.310 | 0.0281 | No |
| 11 | Lgals1 | 2714 | 0.286 | 0.0166 | No |
| 12 | Fzd8 | 2844 | 0.276 | 0.0189 | No |
| 13 | P3h1 | 2907 | 0.270 | 0.0263 | No |
| 14 | Sgcb | 2921 | 0.269 | 0.0377 | No |
| 15 | Mmp2 | 2953 | 0.265 | 0.0474 | No |
| 16 | Cap2 | 2984 | 0.263 | 0.0571 | No |
| 17 | Tgfbr3 | 3096 | 0.256 | 0.0599 | No |
| 18 | Serpinh1 | 3273 | 0.240 | 0.0567 | No |
| 19 | Glipr1 | 3299 | 0.237 | 0.0656 | No |
| 20 | Col4a1 | 3490 | 0.223 | 0.0605 | No |
| 21 | Dst | 3553 | 0.217 | 0.0654 | No |
| 22 | Col1a2 | 3945 | 0.189 | 0.0423 | No |
| 23 | Copa | 4095 | 0.176 | 0.0383 | No |
| 24 | Sntb1 | 4103 | 0.176 | 0.0459 | No |
| 25 | Qsox1 | 4299 | 0.161 | 0.0375 | No |
| 26 | Itga2 | 4496 | 0.147 | 0.0283 | No |
| 27 | Pdgfrb | 4627 | 0.138 | 0.0241 | No |
| 28 | Slit2 | 4847 | 0.123 | 0.0120 | No |
| 29 | Vim | 5016 | 0.112 | 0.0034 | No |
| 30 | Sfrp1 | 5367 | 0.086 | -0.0211 | No |
| 31 | Plod3 | 5412 | 0.083 | -0.0208 | No |
| 32 | Fuca1 | 5767 | 0.058 | -0.0470 | No |
| 33 | Gem | 5849 | 0.051 | -0.0512 | No |
| 34 | Eno2 | 5861 | 0.051 | -0.0498 | No |
| 35 | Sdc4 | 5934 | 0.046 | -0.0535 | No |
| 36 | Plod1 | 5977 | 0.044 | -0.0549 | No |
| 37 | Il15 | 6010 | 0.042 | -0.0556 | No |
| 38 | Tpm1 | 6119 | 0.036 | -0.0627 | No |
| 39 | Col4a2 | 6287 | 0.023 | -0.0753 | No |
| 40 | Vcan | 6549 | 0.006 | -0.0962 | No |
| 41 | Magee1 | 6660 | 0.000 | -0.1052 | No |
| 42 | Abi3bp | 6669 | 0.000 | -0.1059 | No |
| 43 | Basp1 | 6922 | -0.013 | -0.1258 | No |
| 44 | Vegfc | 6945 | -0.014 | -0.1269 | No |
| 45 | Cadm1 | 7118 | -0.027 | -0.1397 | No |
| 46 | Bgn | 7126 | -0.027 | -0.1390 | No |
| 47 | Tpm2 | 7133 | -0.027 | -0.1382 | No |
| 48 | Plaur | 7138 | -0.027 | -0.1373 | No |
| 49 | Gja1 | 7188 | -0.029 | -0.1399 | No |
| 50 | Flna | 7255 | -0.036 | -0.1436 | No |
| 51 | Tnfaip3 | 7275 | -0.038 | -0.1435 | No |
| 52 | Ecm2 | 7298 | -0.039 | -0.1434 | No |
| 53 | Emp3 | 7328 | -0.040 | -0.1439 | No |
| 54 | Mmp14 | 7448 | -0.050 | -0.1513 | No |
| 55 | Bmp1 | 7869 | -0.080 | -0.1818 | No |
| 56 | Mcm7 | 8166 | -0.104 | -0.2011 | No |
| 57 | Itgb3 | 8199 | -0.106 | -0.1988 | No |
| 58 | Colgalt1 | 8437 | -0.125 | -0.2123 | No |
| 59 | Itgav | 8461 | -0.127 | -0.2083 | No |
| 60 | Itga5 | 8794 | -0.157 | -0.2281 | No |
| 61 | Inhba | 9444 | -0.206 | -0.2714 | No |
| 62 | Col16a1 | 9662 | -0.226 | -0.2786 | Yes |
| 63 | Ppib | 9677 | -0.228 | -0.2693 | Yes |
| 64 | Mylk | 9794 | -0.238 | -0.2677 | Yes |
| 65 | Itgb1 | 9942 | -0.255 | -0.2679 | Yes |
| 66 | Igfbp4 | 9948 | -0.256 | -0.2565 | Yes |
| 67 | Calu | 10103 | -0.267 | -0.2567 | Yes |
| 68 | Pmepa1 | 10399 | -0.297 | -0.2670 | Yes |
| 69 | Notch2 | 10528 | -0.314 | -0.2629 | Yes |
| 70 | Jun | 10678 | -0.331 | -0.2597 | Yes |
| 71 | Plod2 | 10771 | -0.344 | -0.2513 | Yes |
| 72 | Tgfbi | 10855 | -0.356 | -0.2417 | Yes |
| 73 | Gpc1 | 11077 | -0.388 | -0.2417 | Yes |
| 74 | Fstl1 | 11147 | -0.393 | -0.2292 | Yes |
| 75 | Sat1 | 11297 | -0.417 | -0.2220 | Yes |
| 76 | Vegfa | 11309 | -0.419 | -0.2036 | Yes |
| 77 | Fas | 11642 | -0.481 | -0.2084 | Yes |
| 78 | Thbs1 | 11746 | -0.504 | -0.1935 | Yes |
| 79 | Cd44 | 11804 | -0.524 | -0.1739 | Yes |
| 80 | Capg | 11857 | -0.537 | -0.1534 | Yes |
| 81 | Fermt2 | 11897 | -0.546 | -0.1313 | Yes |
| 82 | Fap | 11945 | -0.566 | -0.1090 | Yes |
| 83 | Tgm2 | 12135 | -0.657 | -0.0940 | Yes |
| 84 | Wipf1 | 12210 | -0.726 | -0.0665 | Yes |
| 85 | Rhob | 12288 | -0.841 | -0.0339 | Yes |
| 86 | Fbn1 | 12299 | -0.863 | 0.0052 | Yes |