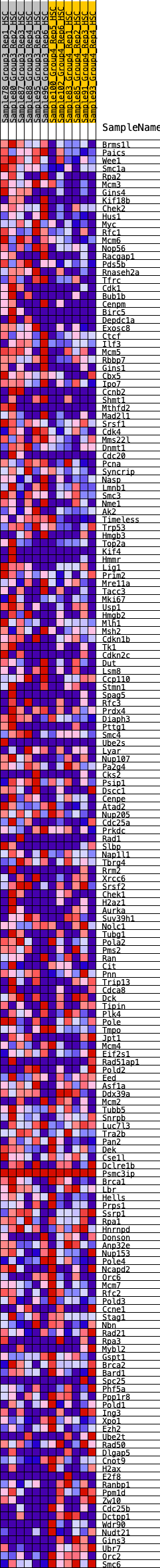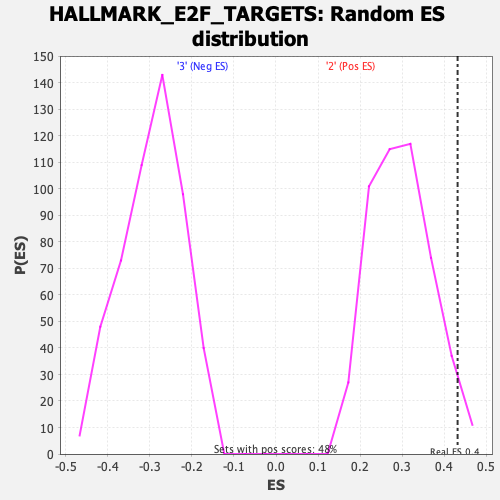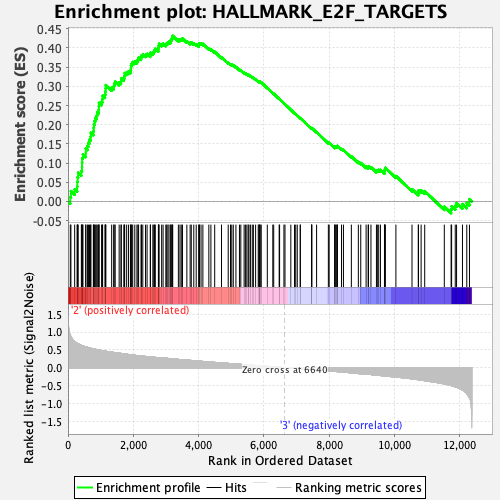
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_E2F_TARGETS |
| Enrichment Score (ES) | 0.43157881 |
| Normalized Enrichment Score (NES) | 1.4487525 |
| Nominal p-value | 0.03526971 |
| FDR q-value | 0.22739154 |
| FWER p-Value | 0.396 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Brms1l | 70 | 0.916 | 0.0118 | Yes |
| 2 | Paics | 89 | 0.872 | 0.0270 | Yes |
| 3 | Wee1 | 204 | 0.729 | 0.0317 | Yes |
| 4 | Smc1a | 281 | 0.684 | 0.0385 | Yes |
| 5 | Rpa2 | 286 | 0.681 | 0.0512 | Yes |
| 6 | Mcm3 | 296 | 0.675 | 0.0634 | Yes |
| 7 | Gins4 | 311 | 0.671 | 0.0751 | Yes |
| 8 | Kif18b | 409 | 0.628 | 0.0792 | Yes |
| 9 | Chek2 | 428 | 0.619 | 0.0896 | Yes |
| 10 | Hus1 | 431 | 0.617 | 0.1013 | Yes |
| 11 | Myc | 435 | 0.615 | 0.1128 | Yes |
| 12 | Rfc1 | 458 | 0.607 | 0.1226 | Yes |
| 13 | Mcm6 | 544 | 0.582 | 0.1268 | Yes |
| 14 | Nop56 | 545 | 0.581 | 0.1379 | Yes |
| 15 | Racgap1 | 595 | 0.571 | 0.1448 | Yes |
| 16 | Pds5b | 626 | 0.562 | 0.1532 | Yes |
| 17 | Rnaseh2a | 658 | 0.557 | 0.1613 | Yes |
| 18 | Tfrc | 693 | 0.546 | 0.1690 | Yes |
| 19 | Cdk1 | 700 | 0.544 | 0.1789 | Yes |
| 20 | Bub1b | 776 | 0.526 | 0.1828 | Yes |
| 21 | Cenpm | 787 | 0.523 | 0.1920 | Yes |
| 22 | Birc5 | 794 | 0.522 | 0.2015 | Yes |
| 23 | Depdc1a | 815 | 0.519 | 0.2098 | Yes |
| 24 | Exosc8 | 837 | 0.516 | 0.2180 | Yes |
| 25 | Ctcf | 880 | 0.508 | 0.2243 | Yes |
| 26 | Ilf3 | 895 | 0.504 | 0.2328 | Yes |
| 27 | Mcm5 | 940 | 0.495 | 0.2386 | Yes |
| 28 | Rbbp7 | 945 | 0.494 | 0.2478 | Yes |
| 29 | Gins1 | 949 | 0.494 | 0.2570 | Yes |
| 30 | Cbx5 | 1028 | 0.481 | 0.2598 | Yes |
| 31 | Ipo7 | 1054 | 0.475 | 0.2669 | Yes |
| 32 | Ccnb2 | 1060 | 0.475 | 0.2755 | Yes |
| 33 | Shmt1 | 1133 | 0.463 | 0.2785 | Yes |
| 34 | Mthfd2 | 1144 | 0.462 | 0.2865 | Yes |
| 35 | Mad2l1 | 1152 | 0.461 | 0.2948 | Yes |
| 36 | Srsf1 | 1157 | 0.460 | 0.3033 | Yes |
| 37 | Cdk4 | 1337 | 0.436 | 0.2969 | Yes |
| 38 | Mms22l | 1399 | 0.427 | 0.3001 | Yes |
| 39 | Dnmt1 | 1413 | 0.425 | 0.3072 | Yes |
| 40 | Cdc20 | 1445 | 0.418 | 0.3127 | Yes |
| 41 | Pcna | 1566 | 0.406 | 0.3106 | Yes |
| 42 | Syncrip | 1623 | 0.400 | 0.3137 | Yes |
| 43 | Nasp | 1631 | 0.398 | 0.3207 | Yes |
| 44 | Lmnb1 | 1709 | 0.388 | 0.3218 | Yes |
| 45 | Smc3 | 1729 | 0.385 | 0.3276 | Yes |
| 46 | Nme1 | 1735 | 0.384 | 0.3346 | Yes |
| 47 | Ak2 | 1796 | 0.376 | 0.3369 | Yes |
| 48 | Timeless | 1854 | 0.368 | 0.3392 | Yes |
| 49 | Trp53 | 1912 | 0.360 | 0.3414 | Yes |
| 50 | Hmgb3 | 1926 | 0.359 | 0.3472 | Yes |
| 51 | Top2a | 1929 | 0.359 | 0.3540 | Yes |
| 52 | Kif4 | 1951 | 0.357 | 0.3591 | Yes |
| 53 | Hmmr | 1981 | 0.353 | 0.3635 | Yes |
| 54 | Lig1 | 2045 | 0.347 | 0.3649 | Yes |
| 55 | Prim2 | 2110 | 0.339 | 0.3662 | Yes |
| 56 | Mre11a | 2141 | 0.336 | 0.3702 | Yes |
| 57 | Tacc3 | 2161 | 0.335 | 0.3750 | Yes |
| 58 | Mki67 | 2236 | 0.327 | 0.3752 | Yes |
| 59 | Usp1 | 2250 | 0.326 | 0.3804 | Yes |
| 60 | Hmgb2 | 2290 | 0.322 | 0.3833 | Yes |
| 61 | Mlh1 | 2374 | 0.314 | 0.3825 | Yes |
| 62 | Msh2 | 2421 | 0.308 | 0.3847 | Yes |
| 63 | Cdkn1b | 2518 | 0.301 | 0.3826 | Yes |
| 64 | Tk1 | 2526 | 0.300 | 0.3877 | Yes |
| 65 | Cdkn2c | 2598 | 0.294 | 0.3876 | Yes |
| 66 | Dut | 2627 | 0.292 | 0.3908 | Yes |
| 67 | Lsm8 | 2649 | 0.290 | 0.3947 | Yes |
| 68 | Ccp110 | 2673 | 0.288 | 0.3983 | Yes |
| 69 | Stmn1 | 2768 | 0.282 | 0.3960 | Yes |
| 70 | Spag5 | 2770 | 0.282 | 0.4013 | Yes |
| 71 | Rfc3 | 2779 | 0.282 | 0.4061 | Yes |
| 72 | Prdx4 | 2786 | 0.281 | 0.4110 | Yes |
| 73 | Diaph3 | 2867 | 0.275 | 0.4096 | Yes |
| 74 | Pttg1 | 2911 | 0.270 | 0.4113 | Yes |
| 75 | Smc4 | 2995 | 0.262 | 0.4095 | Yes |
| 76 | Ube2s | 3022 | 0.261 | 0.4124 | Yes |
| 77 | Lyar | 3058 | 0.259 | 0.4145 | Yes |
| 78 | Nup107 | 3093 | 0.256 | 0.4166 | Yes |
| 79 | Pa2g4 | 3143 | 0.251 | 0.4174 | Yes |
| 80 | Cks2 | 3153 | 0.251 | 0.4214 | Yes |
| 81 | Psip1 | 3178 | 0.249 | 0.4242 | Yes |
| 82 | Dscc1 | 3194 | 0.248 | 0.4278 | Yes |
| 83 | Cenpe | 3206 | 0.246 | 0.4316 | Yes |
| 84 | Atad2 | 3379 | 0.232 | 0.4219 | No |
| 85 | Nup205 | 3428 | 0.226 | 0.4223 | No |
| 86 | Cdc25a | 3468 | 0.225 | 0.4234 | No |
| 87 | Prkdc | 3506 | 0.221 | 0.4246 | No |
| 88 | Rad1 | 3639 | 0.209 | 0.4178 | No |
| 89 | Slbp | 3739 | 0.204 | 0.4136 | No |
| 90 | Nap1l1 | 3774 | 0.202 | 0.4146 | No |
| 91 | Tbrg4 | 3851 | 0.195 | 0.4121 | No |
| 92 | Rrm2 | 3925 | 0.190 | 0.4098 | No |
| 93 | Xrcc6 | 4005 | 0.184 | 0.4068 | No |
| 94 | Srsf2 | 4008 | 0.184 | 0.4102 | No |
| 95 | Chek1 | 4024 | 0.183 | 0.4125 | No |
| 96 | H2az1 | 4073 | 0.179 | 0.4120 | No |
| 97 | Aurka | 4129 | 0.175 | 0.4108 | No |
| 98 | Suv39h1 | 4312 | 0.161 | 0.3989 | No |
| 99 | Nolc1 | 4375 | 0.156 | 0.3968 | No |
| 100 | Tubg1 | 4491 | 0.148 | 0.3902 | No |
| 101 | Pola2 | 4700 | 0.133 | 0.3757 | No |
| 102 | Pms2 | 4905 | 0.120 | 0.3612 | No |
| 103 | Ran | 4975 | 0.114 | 0.3577 | No |
| 104 | Cit | 5014 | 0.112 | 0.3568 | No |
| 105 | Pnn | 5060 | 0.109 | 0.3552 | No |
| 106 | Trip13 | 5141 | 0.103 | 0.3506 | No |
| 107 | Cdca8 | 5249 | 0.095 | 0.3436 | No |
| 108 | Dck | 5291 | 0.092 | 0.3420 | No |
| 109 | Tipin | 5392 | 0.084 | 0.3354 | No |
| 110 | Plk4 | 5431 | 0.082 | 0.3338 | No |
| 111 | Pole | 5475 | 0.079 | 0.3318 | No |
| 112 | Tmpo | 5517 | 0.076 | 0.3299 | No |
| 113 | Jpt1 | 5540 | 0.075 | 0.3296 | No |
| 114 | Mcm4 | 5585 | 0.071 | 0.3273 | No |
| 115 | Eif2s1 | 5641 | 0.066 | 0.3241 | No |
| 116 | Rad51ap1 | 5675 | 0.064 | 0.3226 | No |
| 117 | Pold2 | 5744 | 0.060 | 0.3182 | No |
| 118 | Eed | 5824 | 0.053 | 0.3127 | No |
| 119 | Asf1a | 5850 | 0.051 | 0.3116 | No |
| 120 | Ddx39a | 5856 | 0.051 | 0.3122 | No |
| 121 | Mcm2 | 5871 | 0.050 | 0.3120 | No |
| 122 | Tubb5 | 5884 | 0.049 | 0.3120 | No |
| 123 | Snrpb | 5916 | 0.047 | 0.3103 | No |
| 124 | Luc7l3 | 6102 | 0.037 | 0.2958 | No |
| 125 | Tra2b | 6270 | 0.025 | 0.2826 | No |
| 126 | Pan2 | 6298 | 0.023 | 0.2808 | No |
| 127 | Dek | 6465 | 0.011 | 0.2674 | No |
| 128 | Cse1l | 6478 | 0.010 | 0.2666 | No |
| 129 | Dclre1b | 6611 | 0.002 | 0.2558 | No |
| 130 | Psmc3ip | 6643 | 0.000 | 0.2533 | No |
| 131 | Brca1 | 6822 | -0.006 | 0.2388 | No |
| 132 | Lbr | 6935 | -0.014 | 0.2299 | No |
| 133 | Hells | 6972 | -0.016 | 0.2272 | No |
| 134 | Prps1 | 7019 | -0.020 | 0.2238 | No |
| 135 | Ssrp1 | 7111 | -0.026 | 0.2168 | No |
| 136 | Rpa1 | 7112 | -0.026 | 0.2173 | No |
| 137 | Hnrnpd | 7455 | -0.050 | 0.1902 | No |
| 138 | Donson | 7460 | -0.051 | 0.1909 | No |
| 139 | Anp32e | 7465 | -0.051 | 0.1915 | No |
| 140 | Nup153 | 7611 | -0.060 | 0.1808 | No |
| 141 | Pole4 | 7971 | -0.088 | 0.1530 | No |
| 142 | Ncapd2 | 7996 | -0.091 | 0.1528 | No |
| 143 | Orc6 | 8165 | -0.104 | 0.1410 | No |
| 144 | Mcm7 | 8166 | -0.104 | 0.1429 | No |
| 145 | Rfc2 | 8196 | -0.106 | 0.1426 | No |
| 146 | Pold3 | 8214 | -0.107 | 0.1433 | No |
| 147 | Ccne1 | 8245 | -0.109 | 0.1429 | No |
| 148 | Stag1 | 8247 | -0.109 | 0.1449 | No |
| 149 | Nbn | 8374 | -0.120 | 0.1369 | No |
| 150 | Rad21 | 8433 | -0.125 | 0.1345 | No |
| 151 | Rpa3 | 8675 | -0.145 | 0.1175 | No |
| 152 | Mybl2 | 8889 | -0.163 | 0.1031 | No |
| 153 | Gspt1 | 8963 | -0.169 | 0.1004 | No |
| 154 | Brca2 | 9127 | -0.182 | 0.0905 | No |
| 155 | Bard1 | 9189 | -0.187 | 0.0891 | No |
| 156 | Spc25 | 9200 | -0.188 | 0.0919 | No |
| 157 | Phf5a | 9275 | -0.191 | 0.0895 | No |
| 158 | Ppp1r8 | 9443 | -0.206 | 0.0797 | No |
| 159 | Pold1 | 9470 | -0.210 | 0.0816 | No |
| 160 | Ing3 | 9512 | -0.214 | 0.0823 | No |
| 161 | Xpo1 | 9566 | -0.219 | 0.0822 | No |
| 162 | Ezh2 | 9689 | -0.229 | 0.0765 | No |
| 163 | Ube2t | 9691 | -0.229 | 0.0808 | No |
| 164 | Rad50 | 9703 | -0.230 | 0.0843 | No |
| 165 | Dlgap5 | 9715 | -0.231 | 0.0879 | No |
| 166 | Cnot9 | 10039 | -0.262 | 0.0664 | No |
| 167 | H2ax | 10532 | -0.314 | 0.0320 | No |
| 168 | E2f8 | 10724 | -0.337 | 0.0228 | No |
| 169 | Ranbp1 | 10730 | -0.338 | 0.0289 | No |
| 170 | Ppm1d | 10811 | -0.350 | 0.0290 | No |
| 171 | Zw10 | 10921 | -0.363 | 0.0270 | No |
| 172 | Cdc25b | 11521 | -0.455 | -0.0134 | No |
| 173 | Dctpp1 | 11729 | -0.500 | -0.0209 | No |
| 174 | Wdr90 | 11745 | -0.504 | -0.0124 | No |
| 175 | Nudt21 | 11856 | -0.537 | -0.0112 | No |
| 176 | Gins3 | 11896 | -0.546 | -0.0039 | No |
| 177 | Ubr7 | 12077 | -0.623 | -0.0068 | No |
| 178 | Orc2 | 12209 | -0.726 | -0.0036 | No |
| 179 | Smc6 | 12291 | -0.844 | 0.0059 | No |