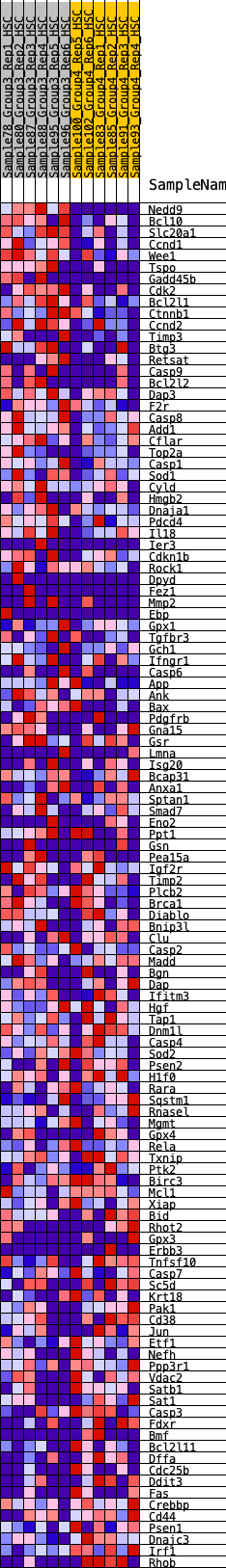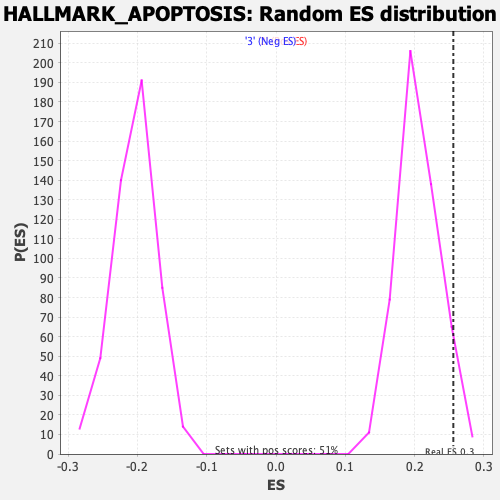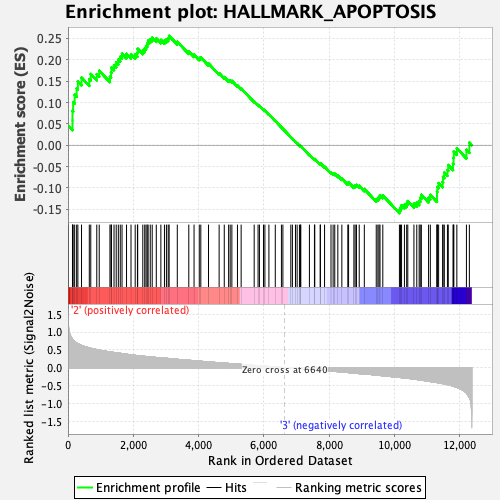
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.25600475 |
| Normalized Enrichment Score (NES) | 1.2417547 |
| Nominal p-value | 0.070866145 |
| FDR q-value | 0.44021636 |
| FWER p-Value | 0.892 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nedd9 | 0 | 1.621 | 0.0464 | Yes |
| 2 | Bcl10 | 137 | 0.790 | 0.0579 | Yes |
| 3 | Slc20a1 | 138 | 0.790 | 0.0805 | Yes |
| 4 | Ccnd1 | 158 | 0.770 | 0.1010 | Yes |
| 5 | Wee1 | 204 | 0.729 | 0.1181 | Yes |
| 6 | Tspo | 265 | 0.693 | 0.1331 | Yes |
| 7 | Gadd45b | 302 | 0.672 | 0.1494 | Yes |
| 8 | Cdk2 | 414 | 0.625 | 0.1582 | Yes |
| 9 | Bcl2l1 | 652 | 0.558 | 0.1548 | Yes |
| 10 | Ctnnb1 | 697 | 0.545 | 0.1668 | Yes |
| 11 | Ccnd2 | 881 | 0.508 | 0.1663 | Yes |
| 12 | Timp3 | 956 | 0.493 | 0.1744 | Yes |
| 13 | Btg3 | 1284 | 0.443 | 0.1604 | Yes |
| 14 | Retsat | 1318 | 0.439 | 0.1702 | Yes |
| 15 | Casp9 | 1331 | 0.437 | 0.1818 | Yes |
| 16 | Bcl2l2 | 1409 | 0.425 | 0.1876 | Yes |
| 17 | Dap3 | 1476 | 0.416 | 0.1941 | Yes |
| 18 | F2r | 1541 | 0.409 | 0.2006 | Yes |
| 19 | Casp8 | 1600 | 0.401 | 0.2074 | Yes |
| 20 | Add1 | 1654 | 0.395 | 0.2143 | Yes |
| 21 | Cflar | 1792 | 0.377 | 0.2139 | Yes |
| 22 | Top2a | 1929 | 0.359 | 0.2131 | Yes |
| 23 | Casp1 | 2057 | 0.346 | 0.2126 | Yes |
| 24 | Sod1 | 2126 | 0.338 | 0.2167 | Yes |
| 25 | Cyld | 2134 | 0.337 | 0.2258 | Yes |
| 26 | Hmgb2 | 2290 | 0.322 | 0.2223 | Yes |
| 27 | Dnaja1 | 2349 | 0.317 | 0.2267 | Yes |
| 28 | Pdcd4 | 2389 | 0.312 | 0.2324 | Yes |
| 29 | Il18 | 2425 | 0.308 | 0.2384 | Yes |
| 30 | Ier3 | 2453 | 0.305 | 0.2449 | Yes |
| 31 | Cdkn1b | 2518 | 0.301 | 0.2483 | Yes |
| 32 | Rock1 | 2578 | 0.295 | 0.2519 | Yes |
| 33 | Dpyd | 2703 | 0.286 | 0.2500 | Yes |
| 34 | Fez1 | 2843 | 0.276 | 0.2465 | Yes |
| 35 | Mmp2 | 2953 | 0.265 | 0.2452 | Yes |
| 36 | Ebp | 3013 | 0.261 | 0.2479 | Yes |
| 37 | Gpx1 | 3067 | 0.259 | 0.2510 | Yes |
| 38 | Tgfbr3 | 3096 | 0.256 | 0.2560 | Yes |
| 39 | Gch1 | 3346 | 0.234 | 0.2424 | No |
| 40 | Ifngr1 | 3698 | 0.206 | 0.2196 | No |
| 41 | Casp6 | 3859 | 0.195 | 0.2121 | No |
| 42 | App | 4019 | 0.183 | 0.2044 | No |
| 43 | Ank | 4066 | 0.179 | 0.2057 | No |
| 44 | Bax | 4302 | 0.161 | 0.1912 | No |
| 45 | Pdgfrb | 4627 | 0.138 | 0.1686 | No |
| 46 | Gna15 | 4784 | 0.127 | 0.1595 | No |
| 47 | Gsr | 4918 | 0.119 | 0.1521 | No |
| 48 | Lmna | 4965 | 0.115 | 0.1516 | No |
| 49 | Isg20 | 5017 | 0.112 | 0.1506 | No |
| 50 | Bcap31 | 5188 | 0.100 | 0.1396 | No |
| 51 | Anxa1 | 5301 | 0.091 | 0.1330 | No |
| 52 | Sptan1 | 5700 | 0.063 | 0.1023 | No |
| 53 | Smad7 | 5819 | 0.054 | 0.0942 | No |
| 54 | Eno2 | 5861 | 0.051 | 0.0923 | No |
| 55 | Ppt1 | 5985 | 0.043 | 0.0835 | No |
| 56 | Gsn | 6028 | 0.041 | 0.0813 | No |
| 57 | Pea15a | 6151 | 0.034 | 0.0723 | No |
| 58 | Igf2r | 6346 | 0.019 | 0.0570 | No |
| 59 | Timp2 | 6534 | 0.007 | 0.0419 | No |
| 60 | Plcb2 | 6579 | 0.004 | 0.0384 | No |
| 61 | Brca1 | 6822 | -0.006 | 0.0188 | No |
| 62 | Diablo | 6870 | -0.009 | 0.0153 | No |
| 63 | Bnip3l | 6964 | -0.016 | 0.0081 | No |
| 64 | Clu | 7015 | -0.020 | 0.0046 | No |
| 65 | Casp2 | 7086 | -0.024 | -0.0004 | No |
| 66 | Madd | 7120 | -0.027 | -0.0024 | No |
| 67 | Bgn | 7126 | -0.027 | -0.0020 | No |
| 68 | Dap | 7391 | -0.046 | -0.0222 | No |
| 69 | Ifitm3 | 7545 | -0.056 | -0.0331 | No |
| 70 | Hgf | 7555 | -0.057 | -0.0322 | No |
| 71 | Tap1 | 7723 | -0.069 | -0.0439 | No |
| 72 | Dnm1l | 7726 | -0.070 | -0.0421 | No |
| 73 | Casp4 | 7855 | -0.079 | -0.0503 | No |
| 74 | Sod2 | 8052 | -0.096 | -0.0635 | No |
| 75 | Psen2 | 8117 | -0.099 | -0.0659 | No |
| 76 | H1f0 | 8167 | -0.104 | -0.0669 | No |
| 77 | Rara | 8261 | -0.110 | -0.0714 | No |
| 78 | Sqstm1 | 8384 | -0.121 | -0.0779 | No |
| 79 | Rnasel | 8567 | -0.134 | -0.0889 | No |
| 80 | Mgmt | 8590 | -0.137 | -0.0868 | No |
| 81 | Gpx4 | 8753 | -0.151 | -0.0957 | No |
| 82 | Rela | 8791 | -0.157 | -0.0942 | No |
| 83 | Txnip | 8832 | -0.160 | -0.0929 | No |
| 84 | Ptk2 | 8916 | -0.165 | -0.0950 | No |
| 85 | Birc3 | 9072 | -0.176 | -0.1026 | No |
| 86 | Mcl1 | 9437 | -0.206 | -0.1264 | No |
| 87 | Xiap | 9487 | -0.212 | -0.1244 | No |
| 88 | Bid | 9522 | -0.216 | -0.1210 | No |
| 89 | Rhot2 | 9552 | -0.218 | -0.1171 | No |
| 90 | Gpx3 | 9638 | -0.224 | -0.1176 | No |
| 91 | Erbb3 | 10147 | -0.274 | -0.1513 | No |
| 92 | Tnfsf10 | 10179 | -0.275 | -0.1460 | No |
| 93 | Casp7 | 10212 | -0.278 | -0.1406 | No |
| 94 | Sc5d | 10300 | -0.287 | -0.1395 | No |
| 95 | Krt18 | 10369 | -0.294 | -0.1366 | No |
| 96 | Pak1 | 10403 | -0.297 | -0.1308 | No |
| 97 | Cd38 | 10591 | -0.320 | -0.1370 | No |
| 98 | Jun | 10678 | -0.331 | -0.1345 | No |
| 99 | Etf1 | 10751 | -0.341 | -0.1306 | No |
| 100 | Nefh | 10784 | -0.346 | -0.1234 | No |
| 101 | Ppp3r1 | 10818 | -0.351 | -0.1160 | No |
| 102 | Vdac2 | 11039 | -0.382 | -0.1230 | No |
| 103 | Satb1 | 11093 | -0.388 | -0.1162 | No |
| 104 | Sat1 | 11297 | -0.417 | -0.1209 | No |
| 105 | Casp3 | 11298 | -0.417 | -0.1090 | No |
| 106 | Fdxr | 11308 | -0.419 | -0.0977 | No |
| 107 | Bmf | 11347 | -0.424 | -0.0887 | No |
| 108 | Bcl2l11 | 11470 | -0.445 | -0.0859 | No |
| 109 | Dffa | 11483 | -0.448 | -0.0741 | No |
| 110 | Cdc25b | 11521 | -0.455 | -0.0641 | No |
| 111 | Ddit3 | 11617 | -0.474 | -0.0583 | No |
| 112 | Fas | 11642 | -0.481 | -0.0465 | No |
| 113 | Crebbp | 11788 | -0.518 | -0.0435 | No |
| 114 | Cd44 | 11804 | -0.524 | -0.0298 | No |
| 115 | Psen1 | 11812 | -0.526 | -0.0153 | No |
| 116 | Dnajc3 | 11905 | -0.549 | -0.0071 | No |
| 117 | Irf1 | 12198 | -0.709 | -0.0107 | No |
| 118 | Rhob | 12288 | -0.841 | 0.0061 | No |