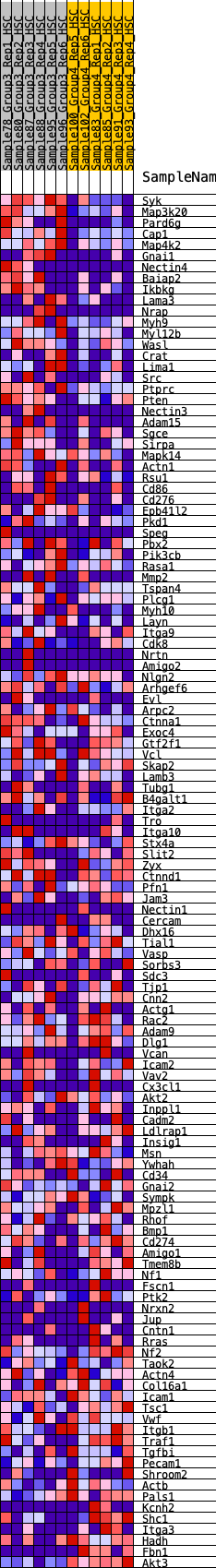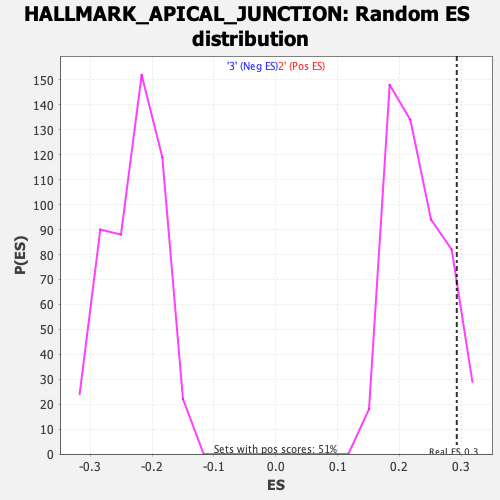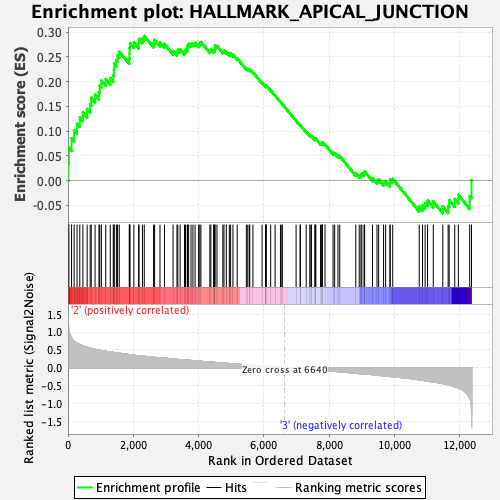
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group3_versus_Group4.HSC_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.29268003 |
| Normalized Enrichment Score (NES) | 1.2782453 |
| Nominal p-value | 0.104950495 |
| FDR q-value | 0.3912975 |
| FWER p-Value | 0.826 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Syk | 13 | 1.219 | 0.0356 | Yes |
| 2 | Map3k20 | 26 | 1.070 | 0.0667 | Yes |
| 3 | Pard6g | 109 | 0.851 | 0.0856 | Yes |
| 4 | Cap1 | 187 | 0.743 | 0.1017 | Yes |
| 5 | Map4k2 | 277 | 0.686 | 0.1150 | Yes |
| 6 | Gnai1 | 361 | 0.650 | 0.1278 | Yes |
| 7 | Nectin4 | 456 | 0.608 | 0.1383 | Yes |
| 8 | Baiap2 | 586 | 0.573 | 0.1450 | Yes |
| 9 | Ikbkg | 679 | 0.550 | 0.1540 | Yes |
| 10 | Lama3 | 711 | 0.540 | 0.1677 | Yes |
| 11 | Nrap | 829 | 0.517 | 0.1737 | Yes |
| 12 | Myh9 | 948 | 0.494 | 0.1789 | Yes |
| 13 | Myl12b | 972 | 0.490 | 0.1918 | Yes |
| 14 | Wasl | 1021 | 0.482 | 0.2023 | Yes |
| 15 | Crat | 1156 | 0.460 | 0.2052 | Yes |
| 16 | Lima1 | 1293 | 0.442 | 0.2074 | Yes |
| 17 | Src | 1386 | 0.429 | 0.2128 | Yes |
| 18 | Ptprc | 1407 | 0.426 | 0.2239 | Yes |
| 19 | Pten | 1412 | 0.425 | 0.2364 | Yes |
| 20 | Nectin3 | 1481 | 0.416 | 0.2433 | Yes |
| 21 | Adam15 | 1517 | 0.412 | 0.2528 | Yes |
| 22 | Sgce | 1569 | 0.405 | 0.2608 | Yes |
| 23 | Sirpa | 1875 | 0.366 | 0.2469 | Yes |
| 24 | Mapk14 | 1883 | 0.365 | 0.2573 | Yes |
| 25 | Actn1 | 1884 | 0.365 | 0.2683 | Yes |
| 26 | Rsu1 | 1899 | 0.363 | 0.2781 | Yes |
| 27 | Cd86 | 2016 | 0.350 | 0.2791 | Yes |
| 28 | Cd276 | 2163 | 0.334 | 0.2772 | Yes |
| 29 | Epb41l2 | 2174 | 0.334 | 0.2864 | Yes |
| 30 | Pkd1 | 2279 | 0.322 | 0.2876 | Yes |
| 31 | Speg | 2335 | 0.318 | 0.2927 | Yes |
| 32 | Pbx2 | 2620 | 0.292 | 0.2783 | No |
| 33 | Pik3cb | 2651 | 0.290 | 0.2845 | No |
| 34 | Rasa1 | 2816 | 0.278 | 0.2795 | No |
| 35 | Mmp2 | 2953 | 0.265 | 0.2763 | No |
| 36 | Tspan4 | 3216 | 0.245 | 0.2623 | No |
| 37 | Plcg1 | 3336 | 0.234 | 0.2596 | No |
| 38 | Myh10 | 3363 | 0.233 | 0.2645 | No |
| 39 | Layn | 3433 | 0.226 | 0.2656 | No |
| 40 | Itga9 | 3566 | 0.216 | 0.2613 | No |
| 41 | Cdk8 | 3598 | 0.212 | 0.2652 | No |
| 42 | Nrtn | 3654 | 0.209 | 0.2670 | No |
| 43 | Amigo2 | 3656 | 0.209 | 0.2732 | No |
| 44 | Nlgn2 | 3692 | 0.208 | 0.2765 | No |
| 45 | Arhgef6 | 3766 | 0.203 | 0.2767 | No |
| 46 | Evl | 3824 | 0.197 | 0.2779 | No |
| 47 | Arpc2 | 3890 | 0.192 | 0.2784 | No |
| 48 | Ctnna1 | 3998 | 0.185 | 0.2752 | No |
| 49 | Exoc4 | 4035 | 0.182 | 0.2777 | No |
| 50 | Gtf2f1 | 4069 | 0.179 | 0.2804 | No |
| 51 | Vcl | 4343 | 0.158 | 0.2629 | No |
| 52 | Skap2 | 4373 | 0.156 | 0.2652 | No |
| 53 | Lamb3 | 4457 | 0.150 | 0.2629 | No |
| 54 | Tubg1 | 4491 | 0.148 | 0.2646 | No |
| 55 | B4galt1 | 4495 | 0.147 | 0.2688 | No |
| 56 | Itga2 | 4496 | 0.147 | 0.2733 | No |
| 57 | Tro | 4561 | 0.143 | 0.2723 | No |
| 58 | Itga10 | 4741 | 0.130 | 0.2616 | No |
| 59 | Stx4a | 4775 | 0.128 | 0.2627 | No |
| 60 | Slit2 | 4847 | 0.123 | 0.2606 | No |
| 61 | Zyx | 4939 | 0.117 | 0.2567 | No |
| 62 | Ctnnd1 | 4978 | 0.114 | 0.2570 | No |
| 63 | Pfn1 | 5050 | 0.109 | 0.2545 | No |
| 64 | Jam3 | 5180 | 0.100 | 0.2470 | No |
| 65 | Nectin1 | 5460 | 0.079 | 0.2266 | No |
| 66 | Cercam | 5503 | 0.077 | 0.2255 | No |
| 67 | Dhx16 | 5550 | 0.074 | 0.2239 | No |
| 68 | Tial1 | 5565 | 0.073 | 0.2250 | No |
| 69 | Vasp | 5662 | 0.065 | 0.2191 | No |
| 70 | Sorbs3 | 5941 | 0.046 | 0.1978 | No |
| 71 | Sdc3 | 6055 | 0.041 | 0.1898 | No |
| 72 | Tjp1 | 6056 | 0.040 | 0.1910 | No |
| 73 | Cnn2 | 6057 | 0.040 | 0.1922 | No |
| 74 | Actg1 | 6067 | 0.040 | 0.1927 | No |
| 75 | Rac2 | 6205 | 0.031 | 0.1824 | No |
| 76 | Adam9 | 6341 | 0.020 | 0.1719 | No |
| 77 | Dlg1 | 6505 | 0.008 | 0.1589 | No |
| 78 | Vcan | 6549 | 0.006 | 0.1555 | No |
| 79 | Icam2 | 6559 | 0.005 | 0.1550 | No |
| 80 | Vav2 | 6983 | -0.017 | 0.1209 | No |
| 81 | Cx3cl1 | 7106 | -0.025 | 0.1117 | No |
| 82 | Akt2 | 7114 | -0.026 | 0.1119 | No |
| 83 | Inppl1 | 7292 | -0.039 | 0.0986 | No |
| 84 | Cadm2 | 7403 | -0.047 | 0.0910 | No |
| 85 | Ldlrap1 | 7439 | -0.049 | 0.0896 | No |
| 86 | Insig1 | 7451 | -0.050 | 0.0902 | No |
| 87 | Msn | 7558 | -0.057 | 0.0833 | No |
| 88 | Ywhah | 7570 | -0.058 | 0.0841 | No |
| 89 | Cd34 | 7588 | -0.059 | 0.0845 | No |
| 90 | Gnai2 | 7732 | -0.070 | 0.0750 | No |
| 91 | Sympk | 7767 | -0.073 | 0.0744 | No |
| 92 | Mpzl1 | 7781 | -0.074 | 0.0755 | No |
| 93 | Rhof | 7786 | -0.074 | 0.0774 | No |
| 94 | Bmp1 | 7869 | -0.080 | 0.0731 | No |
| 95 | Cd274 | 8115 | -0.099 | 0.0561 | No |
| 96 | Amigo1 | 8164 | -0.104 | 0.0553 | No |
| 97 | Tmem8b | 8266 | -0.110 | 0.0503 | No |
| 98 | Nf1 | 8320 | -0.115 | 0.0495 | No |
| 99 | Fscn1 | 8811 | -0.158 | 0.0142 | No |
| 100 | Ptk2 | 8916 | -0.165 | 0.0107 | No |
| 101 | Nrxn2 | 8969 | -0.169 | 0.0115 | No |
| 102 | Jup | 8996 | -0.171 | 0.0145 | No |
| 103 | Cntn1 | 9064 | -0.175 | 0.0143 | No |
| 104 | Rras | 9082 | -0.177 | 0.0182 | No |
| 105 | Nf2 | 9322 | -0.196 | 0.0046 | No |
| 106 | Taok2 | 9460 | -0.209 | -0.0003 | No |
| 107 | Actn4 | 9510 | -0.214 | 0.0021 | No |
| 108 | Col16a1 | 9662 | -0.226 | -0.0034 | No |
| 109 | Icam1 | 9725 | -0.232 | -0.0015 | No |
| 110 | Tsc1 | 9857 | -0.245 | -0.0048 | No |
| 111 | Vwf | 9863 | -0.246 | 0.0021 | No |
| 112 | Itgb1 | 9942 | -0.255 | 0.0034 | No |
| 113 | Traf1 | 10753 | -0.342 | -0.0525 | No |
| 114 | Tgfbi | 10855 | -0.356 | -0.0500 | No |
| 115 | Pecam1 | 10933 | -0.365 | -0.0454 | No |
| 116 | Shroom2 | 11009 | -0.378 | -0.0401 | No |
| 117 | Actb | 11183 | -0.401 | -0.0422 | No |
| 118 | Pals1 | 11475 | -0.447 | -0.0526 | No |
| 119 | Kcnh2 | 11638 | -0.479 | -0.0514 | No |
| 120 | Shc1 | 11670 | -0.485 | -0.0394 | No |
| 121 | Itga3 | 11841 | -0.533 | -0.0372 | No |
| 122 | Hadh | 11951 | -0.567 | -0.0291 | No |
| 123 | Fbn1 | 12299 | -0.863 | -0.0315 | No |
| 124 | Akt3 | 12354 | -1.220 | 0.0007 | No |