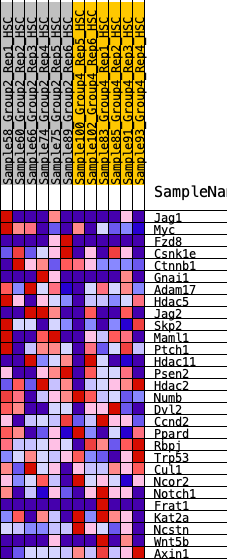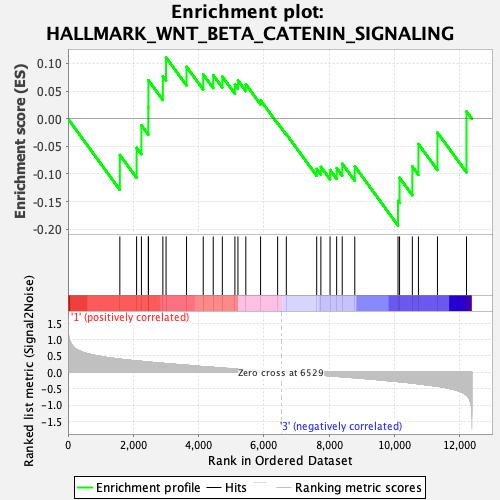
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.19335149 |
| Normalized Enrichment Score (NES) | -0.6903638 |
| Nominal p-value | 0.9241517 |
| FDR q-value | 0.9622199 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Jag1 | 1587 | 0.399 | -0.0654 | No |
| 2 | Myc | 2100 | 0.344 | -0.0524 | No |
| 3 | Fzd8 | 2248 | 0.332 | -0.0117 | No |
| 4 | Csnk1e | 2460 | 0.311 | 0.0205 | No |
| 5 | Ctnnb1 | 2462 | 0.311 | 0.0696 | No |
| 6 | Gnai1 | 2904 | 0.272 | 0.0770 | No |
| 7 | Adam17 | 3002 | 0.264 | 0.1109 | No |
| 8 | Hdac5 | 3629 | 0.214 | 0.0941 | No |
| 9 | Jag2 | 4140 | 0.173 | 0.0802 | No |
| 10 | Skp2 | 4446 | 0.149 | 0.0790 | No |
| 11 | Maml1 | 4725 | 0.125 | 0.0763 | No |
| 12 | Ptch1 | 5110 | 0.104 | 0.0617 | No |
| 13 | Hdac11 | 5203 | 0.096 | 0.0695 | No |
| 14 | Psen2 | 5444 | 0.078 | 0.0624 | No |
| 15 | Hdac2 | 5896 | 0.047 | 0.0333 | No |
| 16 | Numb | 6418 | 0.008 | -0.0077 | No |
| 17 | Dvl2 | 6684 | -0.008 | -0.0279 | No |
| 18 | Ccnd2 | 7614 | -0.079 | -0.0907 | No |
| 19 | Ppard | 7742 | -0.088 | -0.0871 | No |
| 20 | Rbpj | 8023 | -0.108 | -0.0926 | No |
| 21 | Trp53 | 8225 | -0.124 | -0.0893 | No |
| 22 | Cul1 | 8396 | -0.138 | -0.0812 | No |
| 23 | Ncor2 | 8780 | -0.165 | -0.0861 | No |
| 24 | Notch1 | 10104 | -0.283 | -0.1485 | Yes |
| 25 | Frat1 | 10151 | -0.287 | -0.1068 | Yes |
| 26 | Kat2a | 10542 | -0.328 | -0.0864 | Yes |
| 27 | Ncstn | 10729 | -0.352 | -0.0457 | Yes |
| 28 | Wnt5b | 11311 | -0.428 | -0.0250 | Yes |
| 29 | Axin1 | 12200 | -0.695 | 0.0132 | Yes |