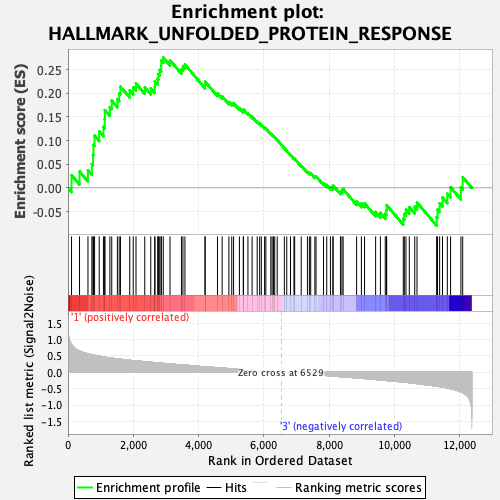
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.2756973 |
| Normalized Enrichment Score (NES) | 1.076639 |
| Nominal p-value | 0.33267328 |
| FDR q-value | 0.6429439 |
| FWER p-Value | 0.981 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nfyb | 108 | 0.847 | 0.0270 | Yes |
| 2 | Yif1a | 353 | 0.651 | 0.0347 | Yes |
| 3 | Nop56 | 611 | 0.561 | 0.0375 | Yes |
| 4 | Exosc10 | 731 | 0.535 | 0.0504 | Yes |
| 5 | Cnot2 | 765 | 0.528 | 0.0701 | Yes |
| 6 | Dnaja4 | 780 | 0.525 | 0.0911 | Yes |
| 7 | Ddx10 | 814 | 0.518 | 0.1104 | Yes |
| 8 | Fkbp14 | 956 | 0.488 | 0.1195 | Yes |
| 9 | Aldh18a1 | 1085 | 0.469 | 0.1289 | Yes |
| 10 | Srprb | 1124 | 0.461 | 0.1453 | Yes |
| 11 | Arfgap1 | 1129 | 0.460 | 0.1645 | Yes |
| 12 | Hsp90b1 | 1282 | 0.440 | 0.1707 | Yes |
| 13 | Edem1 | 1339 | 0.431 | 0.1844 | Yes |
| 14 | Banf1 | 1512 | 0.409 | 0.1876 | Yes |
| 15 | Xbp1 | 1567 | 0.401 | 0.2002 | Yes |
| 16 | Dcp1a | 1606 | 0.397 | 0.2139 | Yes |
| 17 | Eif4a2 | 1890 | 0.365 | 0.2063 | Yes |
| 18 | Psat1 | 1997 | 0.355 | 0.2127 | Yes |
| 19 | Exosc1 | 2082 | 0.347 | 0.2205 | Yes |
| 20 | Lsm4 | 2350 | 0.322 | 0.2123 | Yes |
| 21 | Khsrp | 2534 | 0.306 | 0.2104 | Yes |
| 22 | Imp3 | 2657 | 0.294 | 0.2129 | Yes |
| 23 | Ddit4 | 2660 | 0.294 | 0.2252 | Yes |
| 24 | Mtrex | 2746 | 0.286 | 0.2304 | Yes |
| 25 | Serp1 | 2769 | 0.283 | 0.2405 | Yes |
| 26 | Mthfd2 | 2808 | 0.280 | 0.2493 | Yes |
| 27 | Exoc2 | 2853 | 0.278 | 0.2574 | Yes |
| 28 | Ywhaz | 2855 | 0.278 | 0.2691 | Yes |
| 29 | Pdia5 | 2916 | 0.272 | 0.2757 | Yes |
| 30 | Eif2ak3 | 3123 | 0.253 | 0.2696 | No |
| 31 | Cebpg | 3473 | 0.225 | 0.2507 | No |
| 32 | Hspa5 | 3519 | 0.221 | 0.2564 | No |
| 33 | Dkc1 | 3580 | 0.217 | 0.2606 | No |
| 34 | Parn | 4196 | 0.168 | 0.2176 | No |
| 35 | Srpr | 4198 | 0.168 | 0.2246 | No |
| 36 | Rps14 | 4576 | 0.138 | 0.1998 | No |
| 37 | Bag3 | 4719 | 0.126 | 0.1935 | No |
| 38 | Calr | 4926 | 0.113 | 0.1815 | No |
| 39 | Nop14 | 5013 | 0.110 | 0.1792 | No |
| 40 | Cnot4 | 5069 | 0.107 | 0.1792 | No |
| 41 | Eif4e | 5248 | 0.093 | 0.1686 | No |
| 42 | Tatdn2 | 5365 | 0.084 | 0.1627 | No |
| 43 | Slc30a5 | 5373 | 0.083 | 0.1657 | No |
| 44 | Sec31a | 5509 | 0.074 | 0.1578 | No |
| 45 | Cnot6 | 5636 | 0.065 | 0.1503 | No |
| 46 | Pop4 | 5792 | 0.053 | 0.1399 | No |
| 47 | Edc4 | 5868 | 0.049 | 0.1359 | No |
| 48 | Dctn1 | 5918 | 0.045 | 0.1338 | No |
| 49 | Spcs1 | 6019 | 0.038 | 0.1272 | No |
| 50 | Kif5b | 6057 | 0.035 | 0.1257 | No |
| 51 | Fus | 6210 | 0.023 | 0.1143 | No |
| 52 | Exosc5 | 6249 | 0.020 | 0.1121 | No |
| 53 | Lsm1 | 6290 | 0.017 | 0.1095 | No |
| 54 | Dcp2 | 6329 | 0.015 | 0.1071 | No |
| 55 | Hspa9 | 6405 | 0.009 | 0.1013 | No |
| 56 | Shc1 | 6623 | -0.004 | 0.0838 | No |
| 57 | Gosr2 | 6697 | -0.009 | 0.0782 | No |
| 58 | Cxxc1 | 6811 | -0.017 | 0.0697 | No |
| 59 | Nabp1 | 6918 | -0.025 | 0.0622 | No |
| 60 | Xpot | 6940 | -0.027 | 0.0616 | No |
| 61 | Pdia6 | 7140 | -0.040 | 0.0471 | No |
| 62 | Nfya | 7330 | -0.057 | 0.0340 | No |
| 63 | Eif4a3 | 7390 | -0.061 | 0.0318 | No |
| 64 | Slc7a5 | 7430 | -0.065 | 0.0314 | No |
| 65 | Eif4a1 | 7559 | -0.074 | 0.0241 | No |
| 66 | Nolc1 | 7593 | -0.077 | 0.0246 | No |
| 67 | Atf6 | 7823 | -0.094 | 0.0100 | No |
| 68 | Atf4 | 7921 | -0.101 | 0.0063 | No |
| 69 | Herpud1 | 8035 | -0.109 | 0.0017 | No |
| 70 | Dnajc3 | 8110 | -0.115 | 0.0006 | No |
| 71 | Preb | 8116 | -0.115 | 0.0050 | No |
| 72 | Zbtb17 | 8341 | -0.133 | -0.0076 | No |
| 73 | Ern1 | 8387 | -0.138 | -0.0055 | No |
| 74 | Rrp9 | 8423 | -0.140 | -0.0024 | No |
| 75 | Sdad1 | 8836 | -0.171 | -0.0288 | No |
| 76 | Ssr1 | 8982 | -0.184 | -0.0328 | No |
| 77 | Tspyl2 | 9079 | -0.191 | -0.0325 | No |
| 78 | Eif4ebp1 | 9418 | -0.221 | -0.0507 | No |
| 79 | Npm1 | 9564 | -0.233 | -0.0527 | No |
| 80 | Sec11a | 9713 | -0.248 | -0.0543 | No |
| 81 | Atp6v0d1 | 9752 | -0.252 | -0.0467 | No |
| 82 | Hyou1 | 9756 | -0.252 | -0.0363 | No |
| 83 | Paip1 | 10265 | -0.298 | -0.0651 | No |
| 84 | Exosc2 | 10297 | -0.302 | -0.0548 | No |
| 85 | Ero1a | 10347 | -0.309 | -0.0458 | No |
| 86 | Asns | 10449 | -0.319 | -0.0405 | No |
| 87 | Wipi1 | 10616 | -0.337 | -0.0398 | No |
| 88 | Eif4g1 | 10685 | -0.346 | -0.0307 | No |
| 89 | Eef2 | 11286 | -0.425 | -0.0616 | No |
| 90 | Exosc4 | 11313 | -0.428 | -0.0456 | No |
| 91 | Exosc9 | 11378 | -0.436 | -0.0324 | No |
| 92 | Nhp2 | 11466 | -0.453 | -0.0203 | No |
| 93 | Dnajb9 | 11613 | -0.479 | -0.0119 | No |
| 94 | Eif2s1 | 11713 | -0.504 | 0.0013 | No |
| 95 | Vegfa | 12029 | -0.594 | 0.0007 | No |
| 96 | H2ax | 12084 | -0.624 | 0.0227 | No |

