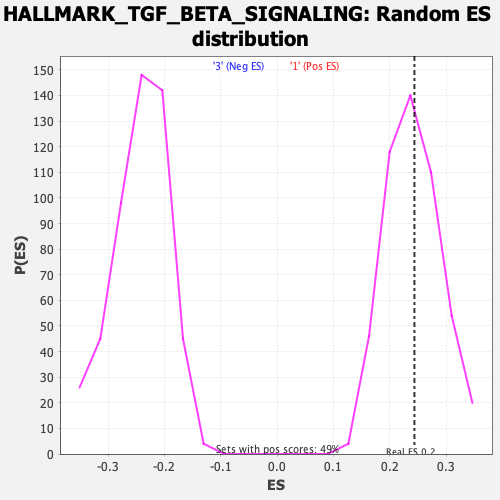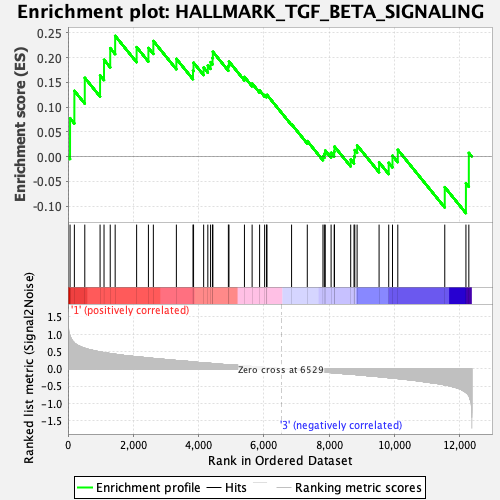
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.24388504 |
| Normalized Enrichment Score (NES) | 1.0143642 |
| Nominal p-value | 0.46341464 |
| FDR q-value | 0.6929619 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acvr1 | 63 | 0.933 | 0.0775 | Yes |
| 2 | Rhoa | 195 | 0.744 | 0.1328 | Yes |
| 3 | Tgfb1 | 514 | 0.589 | 0.1591 | Yes |
| 4 | Smad6 | 982 | 0.485 | 0.1642 | Yes |
| 5 | Ppp1ca | 1103 | 0.464 | 0.1956 | Yes |
| 6 | Tjp1 | 1293 | 0.438 | 0.2191 | Yes |
| 7 | Skil | 1444 | 0.417 | 0.2439 | Yes |
| 8 | Sptbn1 | 2101 | 0.344 | 0.2211 | No |
| 9 | Ctnnb1 | 2462 | 0.311 | 0.2194 | No |
| 10 | Ppp1r15a | 2613 | 0.297 | 0.2336 | No |
| 11 | Ube2d3 | 3318 | 0.238 | 0.1975 | No |
| 12 | Smad3 | 3827 | 0.196 | 0.1736 | No |
| 13 | Ski | 3844 | 0.195 | 0.1896 | No |
| 14 | Smurf1 | 4153 | 0.171 | 0.1798 | No |
| 15 | Cdk9 | 4281 | 0.162 | 0.1838 | No |
| 16 | Map3k7 | 4365 | 0.155 | 0.1908 | No |
| 17 | Smad7 | 4425 | 0.150 | 0.1993 | No |
| 18 | Tgif1 | 4435 | 0.150 | 0.2118 | No |
| 19 | Tgfbr1 | 4910 | 0.114 | 0.1835 | No |
| 20 | Slc20a1 | 4928 | 0.113 | 0.1921 | No |
| 21 | Cdkn1c | 5402 | 0.081 | 0.1609 | No |
| 22 | Hdac1 | 5637 | 0.065 | 0.1477 | No |
| 23 | Apc | 5866 | 0.049 | 0.1335 | No |
| 24 | Ifngr2 | 6020 | 0.038 | 0.1244 | No |
| 25 | Junb | 6079 | 0.034 | 0.1227 | No |
| 26 | Arid4b | 6091 | 0.032 | 0.1247 | No |
| 27 | Bmpr2 | 6845 | -0.019 | 0.0652 | No |
| 28 | Eng | 7326 | -0.056 | 0.0313 | No |
| 29 | Trim33 | 7806 | -0.093 | 0.0006 | No |
| 30 | Xiap | 7849 | -0.096 | 0.0057 | No |
| 31 | Klf10 | 7876 | -0.098 | 0.0122 | No |
| 32 | Smad1 | 8055 | -0.111 | 0.0076 | No |
| 33 | Pmepa1 | 8150 | -0.117 | 0.0104 | No |
| 34 | Fkbp1a | 8159 | -0.118 | 0.0201 | No |
| 35 | Fnta | 8655 | -0.158 | -0.0060 | No |
| 36 | Bmpr1a | 8757 | -0.163 | 0.0002 | No |
| 37 | Ncor2 | 8780 | -0.165 | 0.0131 | No |
| 38 | Smurf2 | 8851 | -0.172 | 0.0226 | No |
| 39 | Bcar3 | 9524 | -0.229 | -0.0116 | No |
| 40 | Furin | 9819 | -0.258 | -0.0126 | No |
| 41 | Wwtr1 | 9935 | -0.268 | 0.0017 | No |
| 42 | Rab31 | 10097 | -0.282 | 0.0137 | No |
| 43 | Thbs1 | 11535 | -0.464 | -0.0619 | No |
| 44 | Ppm1a | 12183 | -0.683 | -0.0539 | No |
| 45 | Hipk2 | 12272 | -0.772 | 0.0074 | No |