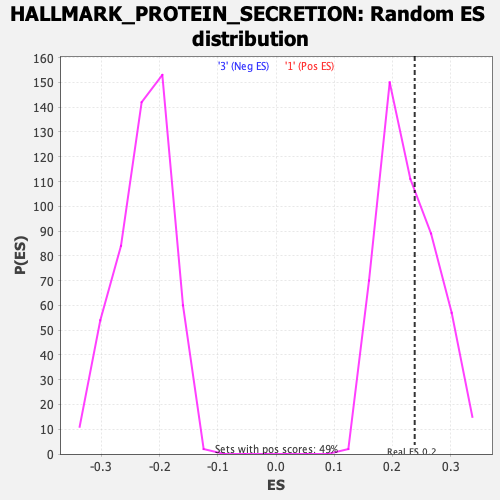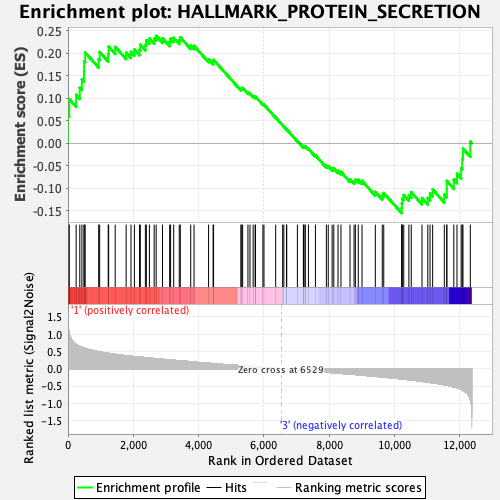
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.23822644 |
| Normalized Enrichment Score (NES) | 1.0443708 |
| Nominal p-value | 0.3987854 |
| FDR q-value | 0.7136152 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cln5 | 1 | 1.631 | 0.0621 | Yes |
| 2 | Abca1 | 39 | 1.009 | 0.0976 | Yes |
| 3 | Copb1 | 249 | 0.701 | 0.1073 | Yes |
| 4 | Vps45 | 361 | 0.648 | 0.1230 | Yes |
| 5 | Yipf6 | 425 | 0.619 | 0.1415 | Yes |
| 6 | Dst | 491 | 0.595 | 0.1589 | Yes |
| 7 | Adam10 | 494 | 0.595 | 0.1814 | Yes |
| 8 | Rab2a | 526 | 0.587 | 0.2013 | Yes |
| 9 | Ap3b1 | 939 | 0.491 | 0.1864 | Yes |
| 10 | Vps4b | 970 | 0.486 | 0.2025 | Yes |
| 11 | Cav2 | 1231 | 0.445 | 0.1983 | Yes |
| 12 | Snap23 | 1245 | 0.444 | 0.2142 | Yes |
| 13 | Stx16 | 1445 | 0.417 | 0.2139 | Yes |
| 14 | Sec24d | 1780 | 0.377 | 0.2011 | Yes |
| 15 | Sgms1 | 1929 | 0.362 | 0.2028 | Yes |
| 16 | Rab9 | 2033 | 0.352 | 0.2078 | Yes |
| 17 | Stam | 2190 | 0.336 | 0.2079 | Yes |
| 18 | Cog2 | 2214 | 0.334 | 0.2188 | Yes |
| 19 | Snx2 | 2369 | 0.320 | 0.2185 | Yes |
| 20 | Dop1a | 2398 | 0.317 | 0.2283 | Yes |
| 21 | Mon2 | 2492 | 0.310 | 0.2325 | Yes |
| 22 | Kif1b | 2638 | 0.296 | 0.2320 | Yes |
| 23 | Rps6ka3 | 2699 | 0.292 | 0.2382 | Yes |
| 24 | Ap1g1 | 2893 | 0.273 | 0.2329 | No |
| 25 | Napg | 3113 | 0.254 | 0.2248 | No |
| 26 | Mapk1 | 3141 | 0.251 | 0.2322 | No |
| 27 | Stx7 | 3236 | 0.245 | 0.2339 | No |
| 28 | Arf1 | 3406 | 0.231 | 0.2289 | No |
| 29 | Zw10 | 3438 | 0.228 | 0.2351 | No |
| 30 | Atp1a1 | 3757 | 0.203 | 0.2169 | No |
| 31 | Arfgap3 | 3859 | 0.194 | 0.2161 | No |
| 32 | Rab5a | 4304 | 0.160 | 0.1860 | No |
| 33 | Sec22b | 4442 | 0.149 | 0.1805 | No |
| 34 | Tmx1 | 4457 | 0.148 | 0.1850 | No |
| 35 | Lamp2 | 5291 | 0.089 | 0.1206 | No |
| 36 | Vamp3 | 5319 | 0.087 | 0.1217 | No |
| 37 | Arfip1 | 5347 | 0.085 | 0.1228 | No |
| 38 | Sec31a | 5509 | 0.074 | 0.1124 | No |
| 39 | Rer1 | 5571 | 0.069 | 0.1101 | No |
| 40 | Ocrl | 5669 | 0.063 | 0.1046 | No |
| 41 | Ppt1 | 5729 | 0.059 | 0.1020 | No |
| 42 | Bet1 | 5740 | 0.058 | 0.1034 | No |
| 43 | Tsg101 | 5967 | 0.042 | 0.0866 | No |
| 44 | Vamp4 | 6000 | 0.039 | 0.0855 | No |
| 45 | Clta | 6358 | 0.013 | 0.0569 | No |
| 46 | Scamp1 | 6572 | -0.000 | 0.0396 | No |
| 47 | Copb2 | 6608 | -0.002 | 0.0368 | No |
| 48 | Stx12 | 6688 | -0.008 | 0.0307 | No |
| 49 | Gosr2 | 6697 | -0.009 | 0.0304 | No |
| 50 | Uso1 | 7023 | -0.031 | 0.0051 | No |
| 51 | Golga4 | 7206 | -0.045 | -0.0080 | No |
| 52 | Tpd52 | 7212 | -0.046 | -0.0067 | No |
| 53 | Arfgef1 | 7256 | -0.049 | -0.0083 | No |
| 54 | Gbf1 | 7262 | -0.050 | -0.0068 | No |
| 55 | Rab22a | 7361 | -0.059 | -0.0125 | No |
| 56 | Cope | 7575 | -0.075 | -0.0270 | No |
| 57 | Napa | 7910 | -0.100 | -0.0504 | No |
| 58 | Arcn1 | 7968 | -0.104 | -0.0511 | No |
| 59 | Anp32e | 8086 | -0.113 | -0.0563 | No |
| 60 | Dnm1l | 8137 | -0.116 | -0.0559 | No |
| 61 | Ap2s1 | 8267 | -0.128 | -0.0616 | No |
| 62 | Bnip3 | 8357 | -0.135 | -0.0637 | No |
| 63 | Ap2m1 | 8634 | -0.157 | -0.0802 | No |
| 64 | Ctsc | 8758 | -0.163 | -0.0840 | No |
| 65 | Tmed10 | 8798 | -0.167 | -0.0808 | No |
| 66 | M6pr | 8887 | -0.175 | -0.0813 | No |
| 67 | Scrn1 | 9002 | -0.187 | -0.0835 | No |
| 68 | Krt18 | 9409 | -0.220 | -0.1081 | No |
| 69 | Cltc | 9622 | -0.239 | -0.1163 | No |
| 70 | Ap2b1 | 9667 | -0.244 | -0.1106 | No |
| 71 | Clcn3 | 10217 | -0.294 | -0.1441 | No |
| 72 | Arfgef2 | 10221 | -0.295 | -0.1331 | No |
| 73 | Scamp3 | 10235 | -0.296 | -0.1229 | No |
| 74 | Atp7a | 10279 | -0.299 | -0.1150 | No |
| 75 | Atp6v1h | 10437 | -0.317 | -0.1157 | No |
| 76 | Ap3s1 | 10509 | -0.326 | -0.1090 | No |
| 77 | Tom1l1 | 10838 | -0.366 | -0.1218 | No |
| 78 | Sod1 | 11016 | -0.388 | -0.1214 | No |
| 79 | Rab14 | 11084 | -0.399 | -0.1117 | No |
| 80 | Ergic3 | 11164 | -0.407 | -0.1026 | No |
| 81 | Ica1 | 11521 | -0.463 | -0.1139 | No |
| 82 | Igf2r | 11597 | -0.476 | -0.1019 | No |
| 83 | Gnas | 11598 | -0.476 | -0.0837 | No |
| 84 | Ykt6 | 11812 | -0.526 | -0.0810 | No |
| 85 | Sspn | 11909 | -0.553 | -0.0677 | No |
| 86 | Lman1 | 12036 | -0.597 | -0.0552 | No |
| 87 | Pam | 12070 | -0.617 | -0.0343 | No |
| 88 | Galc | 12092 | -0.628 | -0.0121 | No |
| 89 | Cd63 | 12317 | -0.894 | 0.0037 | No |