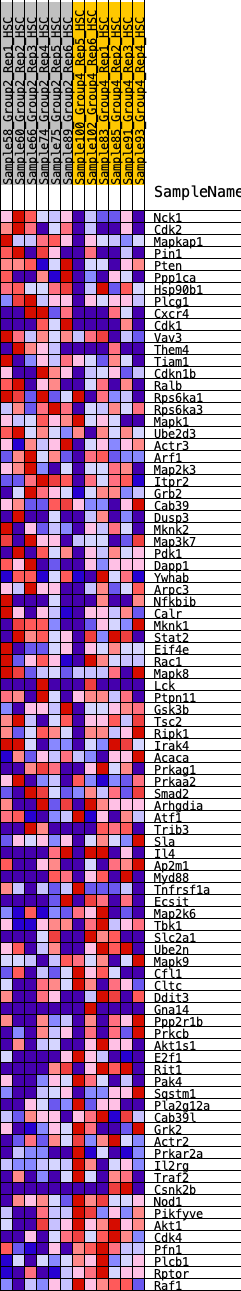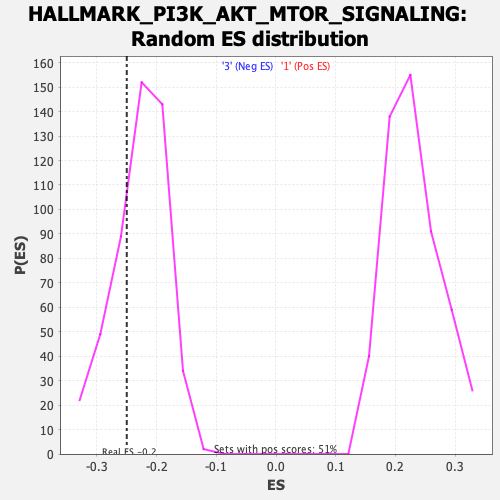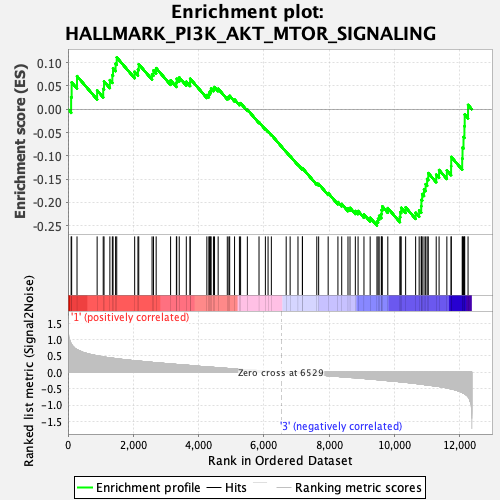
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.24985896 |
| Normalized Enrichment Score (NES) | -1.098284 |
| Nominal p-value | 0.27087575 |
| FDR q-value | 0.8289202 |
| FWER p-Value | 0.953 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nck1 | 96 | 0.872 | 0.0262 | No |
| 2 | Cdk2 | 112 | 0.838 | 0.0576 | No |
| 3 | Mapkap1 | 276 | 0.686 | 0.0711 | No |
| 4 | Pin1 | 892 | 0.501 | 0.0405 | No |
| 5 | Pten | 1078 | 0.470 | 0.0438 | No |
| 6 | Ppp1ca | 1103 | 0.464 | 0.0599 | No |
| 7 | Hsp90b1 | 1282 | 0.440 | 0.0626 | No |
| 8 | Plcg1 | 1358 | 0.429 | 0.0732 | No |
| 9 | Cxcr4 | 1379 | 0.425 | 0.0881 | No |
| 10 | Cdk1 | 1457 | 0.415 | 0.0980 | No |
| 11 | Vav3 | 1492 | 0.412 | 0.1113 | No |
| 12 | Them4 | 2041 | 0.351 | 0.0803 | No |
| 13 | Tiam1 | 2142 | 0.341 | 0.0855 | No |
| 14 | Cdkn1b | 2166 | 0.338 | 0.0968 | No |
| 15 | Ralb | 2574 | 0.301 | 0.0753 | No |
| 16 | Rps6ka1 | 2617 | 0.296 | 0.0835 | No |
| 17 | Rps6ka3 | 2699 | 0.292 | 0.0882 | No |
| 18 | Mapk1 | 3141 | 0.251 | 0.0621 | No |
| 19 | Ube2d3 | 3318 | 0.238 | 0.0570 | No |
| 20 | Actr3 | 3329 | 0.237 | 0.0655 | No |
| 21 | Arf1 | 3406 | 0.231 | 0.0683 | No |
| 22 | Map2k3 | 3622 | 0.215 | 0.0591 | No |
| 23 | Itpr2 | 3738 | 0.205 | 0.0578 | No |
| 24 | Grb2 | 3740 | 0.205 | 0.0657 | No |
| 25 | Cab39 | 4251 | 0.164 | 0.0305 | No |
| 26 | Dusp3 | 4313 | 0.159 | 0.0317 | No |
| 27 | Mknk2 | 4330 | 0.158 | 0.0366 | No |
| 28 | Map3k7 | 4365 | 0.155 | 0.0399 | No |
| 29 | Pdk1 | 4381 | 0.153 | 0.0446 | No |
| 30 | Dapp1 | 4453 | 0.148 | 0.0446 | No |
| 31 | Ywhab | 4486 | 0.146 | 0.0477 | No |
| 32 | Arpc3 | 4597 | 0.137 | 0.0440 | No |
| 33 | Nfkbib | 4877 | 0.116 | 0.0258 | No |
| 34 | Calr | 4926 | 0.113 | 0.0263 | No |
| 35 | Mknk1 | 4950 | 0.112 | 0.0288 | No |
| 36 | Stat2 | 5099 | 0.105 | 0.0209 | No |
| 37 | Eif4e | 5248 | 0.093 | 0.0124 | No |
| 38 | Rac1 | 5284 | 0.090 | 0.0131 | No |
| 39 | Mapk8 | 5491 | 0.075 | -0.0007 | No |
| 40 | Lck | 5848 | 0.050 | -0.0278 | No |
| 41 | Ptpn11 | 6043 | 0.036 | -0.0422 | No |
| 42 | Gsk3b | 6125 | 0.030 | -0.0476 | No |
| 43 | Tsc2 | 6226 | 0.022 | -0.0549 | No |
| 44 | Ripk1 | 6680 | -0.008 | -0.0915 | No |
| 45 | Irak4 | 6801 | -0.017 | -0.1006 | No |
| 46 | Acaca | 7041 | -0.032 | -0.1189 | No |
| 47 | Prkag1 | 7176 | -0.043 | -0.1281 | No |
| 48 | Prkaa2 | 7178 | -0.043 | -0.1265 | No |
| 49 | Smad2 | 7617 | -0.079 | -0.1591 | No |
| 50 | Arhgdia | 7674 | -0.084 | -0.1604 | No |
| 51 | Atf1 | 7965 | -0.104 | -0.1800 | No |
| 52 | Trib3 | 8264 | -0.127 | -0.1993 | No |
| 53 | Sla | 8380 | -0.137 | -0.2033 | No |
| 54 | Il4 | 8572 | -0.153 | -0.2129 | No |
| 55 | Ap2m1 | 8634 | -0.157 | -0.2118 | No |
| 56 | Myd88 | 8797 | -0.166 | -0.2185 | No |
| 57 | Tnfrsf1a | 8881 | -0.174 | -0.2185 | No |
| 58 | Ecsit | 9061 | -0.189 | -0.2257 | No |
| 59 | Map2k6 | 9251 | -0.208 | -0.2330 | No |
| 60 | Tbk1 | 9459 | -0.223 | -0.2412 | Yes |
| 61 | Slc2a1 | 9493 | -0.225 | -0.2351 | Yes |
| 62 | Ube2n | 9526 | -0.229 | -0.2287 | Yes |
| 63 | Mapk9 | 9592 | -0.236 | -0.2248 | Yes |
| 64 | Cfl1 | 9602 | -0.237 | -0.2163 | Yes |
| 65 | Cltc | 9622 | -0.239 | -0.2086 | Yes |
| 66 | Ddit3 | 9791 | -0.255 | -0.2123 | Yes |
| 67 | Gna14 | 10163 | -0.288 | -0.2313 | Yes |
| 68 | Ppp2r1b | 10173 | -0.289 | -0.2208 | Yes |
| 69 | Prkcb | 10201 | -0.291 | -0.2116 | Yes |
| 70 | Akt1s1 | 10337 | -0.307 | -0.2106 | Yes |
| 71 | E2f1 | 10643 | -0.340 | -0.2222 | Yes |
| 72 | Rit1 | 10751 | -0.354 | -0.2172 | Yes |
| 73 | Pak4 | 10814 | -0.362 | -0.2081 | Yes |
| 74 | Sqstm1 | 10820 | -0.362 | -0.1944 | Yes |
| 75 | Pla2g12a | 10844 | -0.366 | -0.1820 | Yes |
| 76 | Cab39l | 10900 | -0.374 | -0.1719 | Yes |
| 77 | Grk2 | 10949 | -0.381 | -0.1609 | Yes |
| 78 | Actr2 | 10995 | -0.386 | -0.1496 | Yes |
| 79 | Prkar2a | 11030 | -0.391 | -0.1371 | Yes |
| 80 | Il2rg | 11273 | -0.424 | -0.1403 | Yes |
| 81 | Traf2 | 11361 | -0.434 | -0.1305 | Yes |
| 82 | Csnk2b | 11599 | -0.477 | -0.1312 | Yes |
| 83 | Nod1 | 11729 | -0.507 | -0.1219 | Yes |
| 84 | Pikfyve | 11733 | -0.508 | -0.1024 | Yes |
| 85 | Akt1 | 12069 | -0.617 | -0.1056 | Yes |
| 86 | Cdk4 | 12079 | -0.621 | -0.0822 | Yes |
| 87 | Pfn1 | 12109 | -0.642 | -0.0595 | Yes |
| 88 | Plcb1 | 12137 | -0.650 | -0.0364 | Yes |
| 89 | Rptor | 12146 | -0.658 | -0.0114 | Yes |
| 90 | Raf1 | 12248 | -0.743 | 0.0094 | Yes |