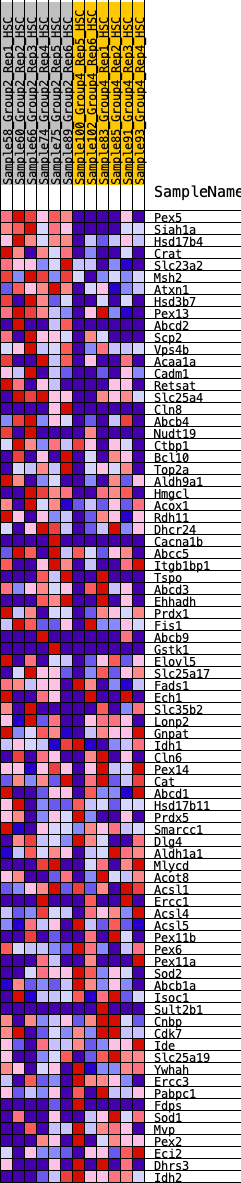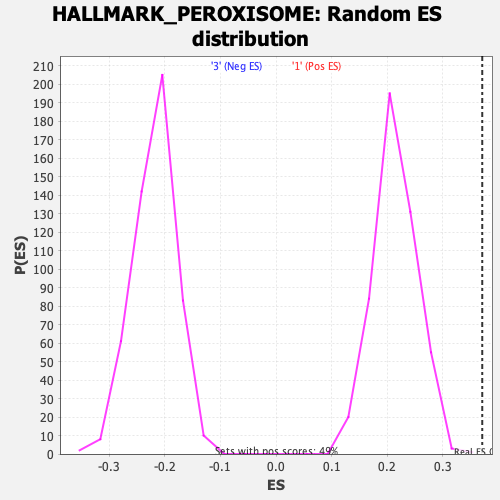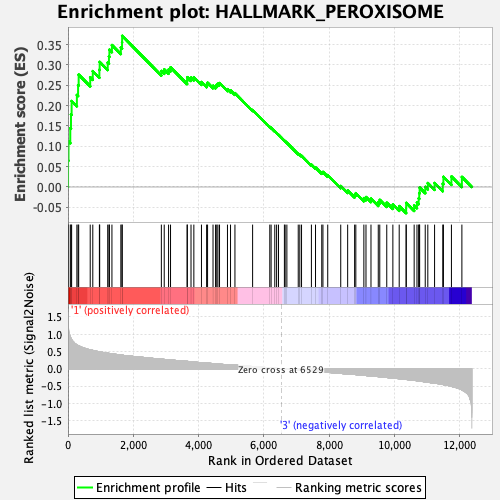
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.371577 |
| Normalized Enrichment Score (NES) | 1.725656 |
| Nominal p-value | 0.0020449897 |
| FDR q-value | 0.026438463 |
| FWER p-Value | 0.027 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 2 | 1.617 | 0.0656 | Yes |
| 2 | Siah1a | 16 | 1.165 | 0.1120 | Yes |
| 3 | Hsd17b4 | 70 | 0.910 | 0.1447 | Yes |
| 4 | Crat | 92 | 0.878 | 0.1787 | Yes |
| 5 | Slc23a2 | 111 | 0.839 | 0.2114 | Yes |
| 6 | Msh2 | 272 | 0.688 | 0.2263 | Yes |
| 7 | Atxn1 | 309 | 0.669 | 0.2506 | Yes |
| 8 | Hsd3b7 | 327 | 0.662 | 0.2762 | Yes |
| 9 | Pex13 | 680 | 0.544 | 0.2697 | Yes |
| 10 | Abcd2 | 759 | 0.530 | 0.2849 | Yes |
| 11 | Scp2 | 961 | 0.488 | 0.2883 | Yes |
| 12 | Vps4b | 970 | 0.486 | 0.3075 | Yes |
| 13 | Acaa1a | 1216 | 0.447 | 0.3057 | Yes |
| 14 | Cadm1 | 1254 | 0.443 | 0.3207 | Yes |
| 15 | Retsat | 1268 | 0.441 | 0.3376 | Yes |
| 16 | Slc25a4 | 1345 | 0.430 | 0.3490 | Yes |
| 17 | Cln8 | 1618 | 0.395 | 0.3429 | Yes |
| 18 | Abcb4 | 1655 | 0.391 | 0.3558 | Yes |
| 19 | Nudt19 | 1658 | 0.391 | 0.3716 | Yes |
| 20 | Ctbp1 | 2858 | 0.277 | 0.2852 | No |
| 21 | Bcl10 | 2946 | 0.269 | 0.2891 | No |
| 22 | Top2a | 3079 | 0.256 | 0.2888 | No |
| 23 | Aldh9a1 | 3139 | 0.252 | 0.2942 | No |
| 24 | Hmgcl | 3647 | 0.213 | 0.2616 | No |
| 25 | Acox1 | 3653 | 0.212 | 0.2698 | No |
| 26 | Rdh11 | 3766 | 0.202 | 0.2690 | No |
| 27 | Dhcr24 | 3853 | 0.195 | 0.2699 | No |
| 28 | Cacna1b | 4086 | 0.174 | 0.2581 | No |
| 29 | Abcc5 | 4244 | 0.165 | 0.2520 | No |
| 30 | Itgb1bp1 | 4273 | 0.162 | 0.2563 | No |
| 31 | Tspo | 4440 | 0.149 | 0.2489 | No |
| 32 | Abcd3 | 4518 | 0.143 | 0.2484 | No |
| 33 | Ehhadh | 4555 | 0.140 | 0.2512 | No |
| 34 | Prdx1 | 4590 | 0.137 | 0.2540 | No |
| 35 | Fis1 | 4639 | 0.133 | 0.2555 | No |
| 36 | Abcb9 | 4882 | 0.115 | 0.2405 | No |
| 37 | Gstk1 | 4976 | 0.110 | 0.2374 | No |
| 38 | Elovl5 | 5111 | 0.104 | 0.2307 | No |
| 39 | Slc25a17 | 5652 | 0.064 | 0.1894 | No |
| 40 | Fads1 | 6178 | 0.026 | 0.1477 | No |
| 41 | Ech1 | 6218 | 0.023 | 0.1454 | No |
| 42 | Slc35b2 | 6331 | 0.015 | 0.1369 | No |
| 43 | Lonp2 | 6387 | 0.010 | 0.1328 | No |
| 44 | Gnpat | 6444 | 0.006 | 0.1285 | No |
| 45 | Idh1 | 6625 | -0.004 | 0.1140 | No |
| 46 | Cln6 | 6648 | -0.005 | 0.1124 | No |
| 47 | Pex14 | 6701 | -0.009 | 0.1086 | No |
| 48 | Cat | 7049 | -0.033 | 0.0817 | No |
| 49 | Abcd1 | 7097 | -0.036 | 0.0793 | No |
| 50 | Hsd17b11 | 7150 | -0.041 | 0.0768 | No |
| 51 | Prdx5 | 7452 | -0.066 | 0.0549 | No |
| 52 | Smarcc1 | 7578 | -0.076 | 0.0479 | No |
| 53 | Dlg4 | 7769 | -0.090 | 0.0361 | No |
| 54 | Aldh1a1 | 7805 | -0.093 | 0.0370 | No |
| 55 | Mlycd | 7952 | -0.103 | 0.0293 | No |
| 56 | Acot8 | 8350 | -0.134 | 0.0024 | No |
| 57 | Acsl1 | 8562 | -0.152 | -0.0086 | No |
| 58 | Ercc1 | 8775 | -0.165 | -0.0191 | No |
| 59 | Acsl4 | 8812 | -0.168 | -0.0152 | No |
| 60 | Acsl5 | 9058 | -0.188 | -0.0275 | No |
| 61 | Pex11b | 9123 | -0.195 | -0.0248 | No |
| 62 | Pex6 | 9276 | -0.211 | -0.0286 | No |
| 63 | Pex11a | 9499 | -0.226 | -0.0374 | No |
| 64 | Sod2 | 9545 | -0.231 | -0.0317 | No |
| 65 | Abcb1a | 9759 | -0.253 | -0.0388 | No |
| 66 | Isoc1 | 9946 | -0.269 | -0.0430 | No |
| 67 | Sult2b1 | 10140 | -0.287 | -0.0470 | No |
| 68 | Cnbp | 10353 | -0.309 | -0.0517 | No |
| 69 | Cdk7 | 10354 | -0.309 | -0.0391 | No |
| 70 | Ide | 10598 | -0.334 | -0.0453 | No |
| 71 | Slc25a19 | 10684 | -0.346 | -0.0382 | No |
| 72 | Ywhah | 10736 | -0.352 | -0.0280 | No |
| 73 | Ercc3 | 10754 | -0.354 | -0.0150 | No |
| 74 | Pabpc1 | 10766 | -0.356 | -0.0014 | No |
| 75 | Fdps | 10937 | -0.380 | 0.0002 | No |
| 76 | Sod1 | 11016 | -0.388 | 0.0097 | No |
| 77 | Mvp | 11223 | -0.415 | 0.0098 | No |
| 78 | Pex2 | 11474 | -0.454 | 0.0079 | No |
| 79 | Eci2 | 11494 | -0.457 | 0.0250 | No |
| 80 | Dhrs3 | 11739 | -0.508 | 0.0258 | No |
| 81 | Idh2 | 12059 | -0.612 | 0.0247 | No |