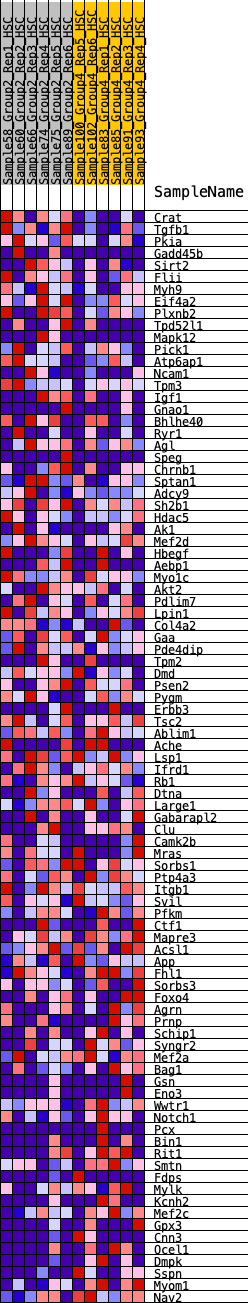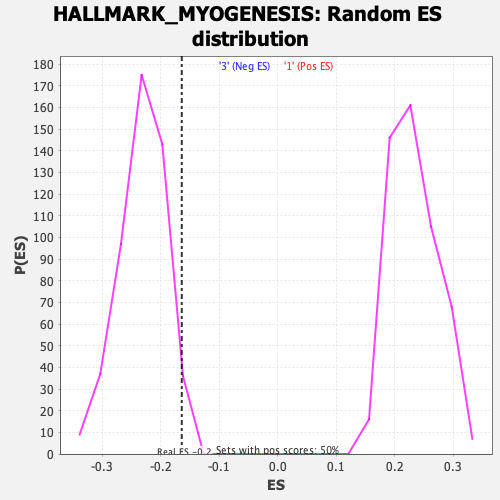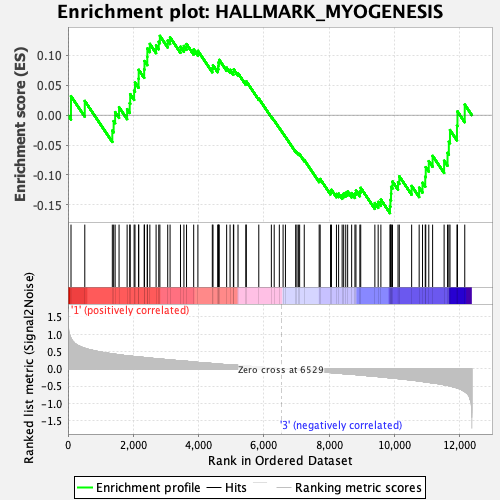
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.1638627 |
| Normalized Enrichment Score (NES) | -0.70726305 |
| Nominal p-value | 0.9839034 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Crat | 92 | 0.878 | 0.0318 | No |
| 2 | Tgfb1 | 514 | 0.589 | 0.0238 | No |
| 3 | Pkia | 1357 | 0.429 | -0.0257 | No |
| 4 | Gadd45b | 1398 | 0.423 | -0.0100 | No |
| 5 | Sirt2 | 1443 | 0.417 | 0.0051 | No |
| 6 | Flii | 1563 | 0.402 | 0.0133 | No |
| 7 | Myh9 | 1810 | 0.374 | 0.0100 | No |
| 8 | Eif4a2 | 1890 | 0.365 | 0.0199 | No |
| 9 | Plxnb2 | 1904 | 0.364 | 0.0351 | No |
| 10 | Tpd52l1 | 2024 | 0.352 | 0.0411 | No |
| 11 | Mapk12 | 2051 | 0.349 | 0.0546 | No |
| 12 | Pick1 | 2161 | 0.339 | 0.0609 | No |
| 13 | Atp6ap1 | 2165 | 0.338 | 0.0758 | No |
| 14 | Ncam1 | 2332 | 0.324 | 0.0767 | No |
| 15 | Tpm3 | 2342 | 0.323 | 0.0904 | No |
| 16 | Igf1 | 2427 | 0.314 | 0.0976 | No |
| 17 | Gnao1 | 2430 | 0.314 | 0.1115 | No |
| 18 | Bhlhe40 | 2506 | 0.308 | 0.1191 | No |
| 19 | Ryr1 | 2697 | 0.292 | 0.1167 | No |
| 20 | Agl | 2779 | 0.283 | 0.1227 | No |
| 21 | Speg | 2813 | 0.280 | 0.1326 | No |
| 22 | Chrnb1 | 3053 | 0.259 | 0.1246 | No |
| 23 | Sptan1 | 3125 | 0.253 | 0.1302 | No |
| 24 | Adcy9 | 3444 | 0.227 | 0.1144 | No |
| 25 | Sh2b1 | 3549 | 0.218 | 0.1157 | No |
| 26 | Hdac5 | 3629 | 0.214 | 0.1189 | No |
| 27 | Ak1 | 3848 | 0.195 | 0.1098 | No |
| 28 | Mef2d | 3977 | 0.182 | 0.1075 | No |
| 29 | Hbegf | 4418 | 0.151 | 0.0784 | No |
| 30 | Aebp1 | 4439 | 0.149 | 0.0834 | No |
| 31 | Myo1c | 4583 | 0.138 | 0.0779 | No |
| 32 | Akt2 | 4604 | 0.136 | 0.0824 | No |
| 33 | Pdlim7 | 4608 | 0.136 | 0.0882 | No |
| 34 | Lpin1 | 4631 | 0.134 | 0.0924 | No |
| 35 | Col4a2 | 4857 | 0.117 | 0.0793 | No |
| 36 | Gaa | 4962 | 0.110 | 0.0758 | No |
| 37 | Pde4dip | 5064 | 0.107 | 0.0723 | No |
| 38 | Tpm2 | 5074 | 0.107 | 0.0764 | No |
| 39 | Dmd | 5205 | 0.096 | 0.0701 | No |
| 40 | Psen2 | 5444 | 0.078 | 0.0542 | No |
| 41 | Pygm | 5459 | 0.077 | 0.0565 | No |
| 42 | Erbb3 | 5841 | 0.050 | 0.0277 | No |
| 43 | Tsc2 | 6226 | 0.022 | -0.0026 | No |
| 44 | Ablim1 | 6313 | 0.016 | -0.0089 | No |
| 45 | Ache | 6470 | 0.004 | -0.0214 | No |
| 46 | Lsp1 | 6585 | -0.001 | -0.0307 | No |
| 47 | Ifrd1 | 6657 | -0.006 | -0.0362 | No |
| 48 | Rb1 | 6970 | -0.029 | -0.0604 | No |
| 49 | Dtna | 7009 | -0.030 | -0.0621 | No |
| 50 | Large1 | 7053 | -0.033 | -0.0641 | No |
| 51 | Gabarapl2 | 7092 | -0.036 | -0.0656 | No |
| 52 | Clu | 7233 | -0.048 | -0.0749 | No |
| 53 | Camk2b | 7690 | -0.085 | -0.1082 | No |
| 54 | Mras | 7720 | -0.087 | -0.1067 | No |
| 55 | Sorbs1 | 8038 | -0.109 | -0.1277 | No |
| 56 | Ptp4a3 | 8066 | -0.112 | -0.1249 | No |
| 57 | Itgb1 | 8219 | -0.123 | -0.1318 | No |
| 58 | Svil | 8285 | -0.129 | -0.1313 | No |
| 59 | Pfkm | 8393 | -0.138 | -0.1338 | No |
| 60 | Ctf1 | 8440 | -0.142 | -0.1312 | No |
| 61 | Mapre3 | 8503 | -0.147 | -0.1297 | No |
| 62 | Acsl1 | 8562 | -0.152 | -0.1277 | No |
| 63 | App | 8684 | -0.161 | -0.1303 | No |
| 64 | Fhl1 | 8785 | -0.166 | -0.1311 | No |
| 65 | Sorbs3 | 8816 | -0.169 | -0.1260 | No |
| 66 | Foxo4 | 8936 | -0.179 | -0.1276 | No |
| 67 | Agrn | 8962 | -0.182 | -0.1215 | No |
| 68 | Prnp | 9395 | -0.219 | -0.1470 | No |
| 69 | Schip1 | 9496 | -0.226 | -0.1450 | No |
| 70 | Syngr2 | 9579 | -0.235 | -0.1412 | No |
| 71 | Mef2a | 9858 | -0.261 | -0.1522 | Yes |
| 72 | Bag1 | 9871 | -0.262 | -0.1415 | Yes |
| 73 | Gsn | 9890 | -0.263 | -0.1312 | Yes |
| 74 | Eno3 | 9895 | -0.263 | -0.1198 | Yes |
| 75 | Wwtr1 | 9935 | -0.268 | -0.1110 | Yes |
| 76 | Notch1 | 10104 | -0.283 | -0.1120 | Yes |
| 77 | Pcx | 10143 | -0.287 | -0.1023 | Yes |
| 78 | Bin1 | 10521 | -0.328 | -0.1184 | Yes |
| 79 | Rit1 | 10751 | -0.354 | -0.1212 | Yes |
| 80 | Smtn | 10854 | -0.368 | -0.1131 | Yes |
| 81 | Fdps | 10937 | -0.380 | -0.1028 | Yes |
| 82 | Mylk | 10953 | -0.382 | -0.0869 | Yes |
| 83 | Kcnh2 | 11046 | -0.393 | -0.0769 | Yes |
| 84 | Mef2c | 11163 | -0.407 | -0.0681 | Yes |
| 85 | Gpx3 | 11515 | -0.461 | -0.0761 | Yes |
| 86 | Cnn3 | 11620 | -0.481 | -0.0631 | Yes |
| 87 | Ocel1 | 11660 | -0.488 | -0.0445 | Yes |
| 88 | Dmpk | 11695 | -0.499 | -0.0249 | Yes |
| 89 | Sspn | 11909 | -0.553 | -0.0175 | Yes |
| 90 | Myom1 | 11924 | -0.558 | 0.0063 | Yes |
| 91 | Nav2 | 12147 | -0.658 | 0.0176 | Yes |