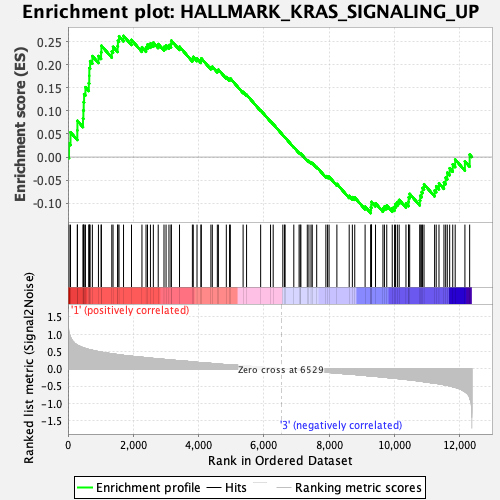
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.2627505 |
| Normalized Enrichment Score (NES) | 1.2749879 |
| Nominal p-value | 0.032258064 |
| FDR q-value | 0.9284381 |
| FWER p-Value | 0.766 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | F13a1 | 32 | 1.036 | 0.0294 | Yes |
| 2 | Sdccag8 | 77 | 0.903 | 0.0538 | Yes |
| 3 | Evi5 | 285 | 0.680 | 0.0579 | Yes |
| 4 | Gadd45g | 289 | 0.678 | 0.0786 | Yes |
| 5 | Spon1 | 459 | 0.605 | 0.0836 | Yes |
| 6 | Crot | 471 | 0.602 | 0.1013 | Yes |
| 7 | Birc3 | 481 | 0.599 | 0.1191 | Yes |
| 8 | Dcbld2 | 493 | 0.595 | 0.1366 | Yes |
| 9 | Il1rl2 | 534 | 0.585 | 0.1514 | Yes |
| 10 | Tmem176a | 637 | 0.554 | 0.1602 | Yes |
| 11 | Cd37 | 651 | 0.550 | 0.1762 | Yes |
| 12 | Klf4 | 653 | 0.550 | 0.1931 | Yes |
| 13 | Csf2ra | 677 | 0.545 | 0.2081 | Yes |
| 14 | Trib1 | 746 | 0.531 | 0.2190 | Yes |
| 15 | St6gal1 | 932 | 0.492 | 0.2191 | Yes |
| 16 | Pdcd1lg2 | 1013 | 0.480 | 0.2274 | Yes |
| 17 | Gucy1a1 | 1023 | 0.479 | 0.2415 | Yes |
| 18 | Ets1 | 1341 | 0.431 | 0.2289 | Yes |
| 19 | Cxcr4 | 1379 | 0.425 | 0.2391 | Yes |
| 20 | Rabgap1l | 1519 | 0.408 | 0.2403 | Yes |
| 21 | Nin | 1528 | 0.407 | 0.2523 | Yes |
| 22 | Il10ra | 1562 | 0.402 | 0.2620 | Yes |
| 23 | Vwa5a | 1700 | 0.385 | 0.2628 | Yes |
| 24 | Usp12 | 1944 | 0.360 | 0.2540 | No |
| 25 | Btc | 2264 | 0.331 | 0.2382 | No |
| 26 | Plaur | 2389 | 0.319 | 0.2380 | No |
| 27 | Bpgm | 2434 | 0.313 | 0.2441 | No |
| 28 | Ephb2 | 2524 | 0.307 | 0.2463 | No |
| 29 | Ppp1r15a | 2613 | 0.297 | 0.2483 | No |
| 30 | Rbm4 | 2762 | 0.284 | 0.2450 | No |
| 31 | Ptcd2 | 2939 | 0.270 | 0.2390 | No |
| 32 | Adam17 | 3002 | 0.264 | 0.2421 | No |
| 33 | Avl9 | 3091 | 0.255 | 0.2428 | No |
| 34 | Gprc5b | 3155 | 0.250 | 0.2454 | No |
| 35 | Ptbp2 | 3163 | 0.249 | 0.2526 | No |
| 36 | Atg10 | 3414 | 0.230 | 0.2393 | No |
| 37 | Mtmr10 | 3808 | 0.199 | 0.2134 | No |
| 38 | Yrdc | 3835 | 0.196 | 0.2173 | No |
| 39 | Btbd3 | 3950 | 0.185 | 0.2137 | No |
| 40 | Gng11 | 4069 | 0.175 | 0.2095 | No |
| 41 | Mmd | 4081 | 0.174 | 0.2140 | No |
| 42 | Ank | 4375 | 0.154 | 0.1948 | No |
| 43 | Hbegf | 4418 | 0.151 | 0.1961 | No |
| 44 | Satb1 | 4572 | 0.138 | 0.1879 | No |
| 45 | Akt2 | 4604 | 0.136 | 0.1895 | No |
| 46 | Tnfrsf1b | 4847 | 0.118 | 0.1734 | No |
| 47 | Lcp1 | 4946 | 0.112 | 0.1689 | No |
| 48 | Ly96 | 4970 | 0.110 | 0.1705 | No |
| 49 | Cbr4 | 5361 | 0.084 | 0.1413 | No |
| 50 | Tor1aip2 | 5467 | 0.077 | 0.1351 | No |
| 51 | Jup | 5898 | 0.047 | 0.1014 | No |
| 52 | Adgra2 | 6201 | 0.024 | 0.0775 | No |
| 53 | Adgrl4 | 6282 | 0.017 | 0.0715 | No |
| 54 | Car2 | 6577 | -0.001 | 0.0476 | No |
| 55 | Wdr33 | 6627 | -0.004 | 0.0437 | No |
| 56 | Map3k1 | 6647 | -0.005 | 0.0423 | No |
| 57 | Map7 | 6913 | -0.024 | 0.0214 | No |
| 58 | Dock2 | 7075 | -0.034 | 0.0094 | No |
| 59 | Itga2 | 7109 | -0.038 | 0.0078 | No |
| 60 | Dnmbp | 7130 | -0.040 | 0.0074 | No |
| 61 | Eng | 7326 | -0.056 | -0.0067 | No |
| 62 | Inhba | 7376 | -0.061 | -0.0089 | No |
| 63 | Ccser2 | 7440 | -0.065 | -0.0120 | No |
| 64 | Epb41l3 | 7478 | -0.068 | -0.0129 | No |
| 65 | Ccnd2 | 7614 | -0.079 | -0.0215 | No |
| 66 | Tmem176b | 7895 | -0.099 | -0.0413 | No |
| 67 | Tspan13 | 7947 | -0.102 | -0.0422 | No |
| 68 | Strn | 7993 | -0.106 | -0.0426 | No |
| 69 | Pecam1 | 8232 | -0.124 | -0.0582 | No |
| 70 | Psmb8 | 8609 | -0.155 | -0.0841 | No |
| 71 | Lat2 | 8706 | -0.161 | -0.0869 | No |
| 72 | Zfp639 | 8779 | -0.165 | -0.0877 | No |
| 73 | Cdadc1 | 9096 | -0.193 | -0.1075 | No |
| 74 | Irf8 | 9270 | -0.210 | -0.1151 | No |
| 75 | Itgb2 | 9273 | -0.211 | -0.1088 | No |
| 76 | Trib2 | 9291 | -0.212 | -0.1036 | No |
| 77 | Etv5 | 9293 | -0.212 | -0.0972 | No |
| 78 | Mycn | 9415 | -0.220 | -0.1002 | No |
| 79 | Glrx | 9642 | -0.241 | -0.1112 | No |
| 80 | Ammecr1 | 9688 | -0.246 | -0.1073 | No |
| 81 | Abcb1a | 9759 | -0.253 | -0.1052 | No |
| 82 | Tnfaip3 | 9927 | -0.267 | -0.1105 | No |
| 83 | Peg3 | 9998 | -0.274 | -0.1078 | No |
| 84 | Gypc | 10028 | -0.276 | -0.1016 | No |
| 85 | Kif5c | 10084 | -0.282 | -0.0974 | No |
| 86 | Tfpi | 10139 | -0.287 | -0.0929 | No |
| 87 | Ero1a | 10347 | -0.309 | -0.1002 | No |
| 88 | Tmem158 | 10425 | -0.316 | -0.0968 | No |
| 89 | Zfp277 | 10430 | -0.316 | -0.0873 | No |
| 90 | Hdac9 | 10461 | -0.320 | -0.0799 | No |
| 91 | Map4k1 | 10770 | -0.356 | -0.0940 | No |
| 92 | Fuca1 | 10783 | -0.358 | -0.0839 | No |
| 93 | Ctss | 10827 | -0.364 | -0.0761 | No |
| 94 | Gfpt2 | 10855 | -0.368 | -0.0670 | No |
| 95 | Cab39l | 10900 | -0.374 | -0.0590 | No |
| 96 | Laptm5 | 11224 | -0.415 | -0.0725 | No |
| 97 | Il2rg | 11273 | -0.424 | -0.0633 | No |
| 98 | Dusp6 | 11358 | -0.433 | -0.0567 | No |
| 99 | Igf2 | 11510 | -0.460 | -0.0548 | No |
| 100 | Ikzf1 | 11563 | -0.468 | -0.0446 | No |
| 101 | Cbl | 11612 | -0.479 | -0.0337 | No |
| 102 | Prelid3b | 11685 | -0.495 | -0.0243 | No |
| 103 | Traf1 | 11779 | -0.518 | -0.0158 | No |
| 104 | Kcnn4 | 11854 | -0.536 | -0.0053 | No |
| 105 | Spry2 | 12152 | -0.660 | -0.0091 | No |
| 106 | Fbxo4 | 12297 | -0.848 | 0.0054 | No |

