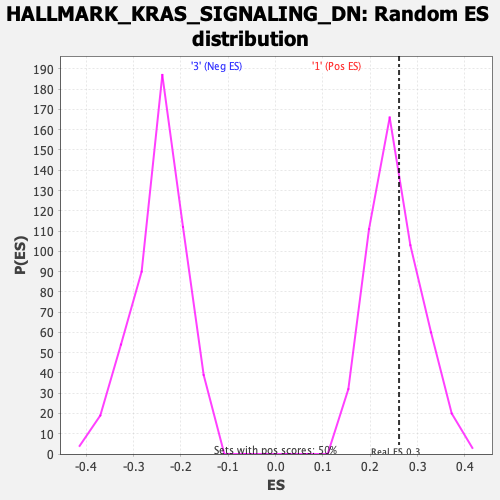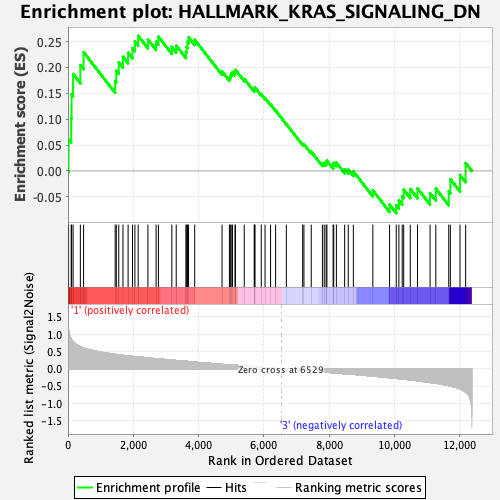
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.26086947 |
| Normalized Enrichment Score (NES) | 1.0372697 |
| Nominal p-value | 0.3858586 |
| FDR q-value | 0.6904205 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Camk1d | 18 | 1.149 | 0.0615 | Yes |
| 2 | Myo15a | 98 | 0.862 | 0.1024 | Yes |
| 3 | Slc25a23 | 109 | 0.845 | 0.1479 | Yes |
| 4 | Ypel1 | 156 | 0.786 | 0.1873 | Yes |
| 5 | Tent5c | 378 | 0.640 | 0.2044 | Yes |
| 6 | Ccdc106 | 477 | 0.601 | 0.2294 | Yes |
| 7 | Skil | 1444 | 0.417 | 0.1737 | Yes |
| 8 | Edar | 1479 | 0.413 | 0.1936 | Yes |
| 9 | Kcnd1 | 1554 | 0.403 | 0.2098 | Yes |
| 10 | Egf | 1683 | 0.388 | 0.2206 | Yes |
| 11 | Fggy | 1840 | 0.372 | 0.2283 | Yes |
| 12 | Copz2 | 1974 | 0.358 | 0.2371 | Yes |
| 13 | Slc16a7 | 2048 | 0.350 | 0.2504 | Yes |
| 14 | Ntf3 | 2149 | 0.340 | 0.2609 | Yes |
| 15 | Thnsl2 | 2447 | 0.312 | 0.2538 | No |
| 16 | Ryr1 | 2697 | 0.292 | 0.2496 | No |
| 17 | Gpr19 | 2768 | 0.283 | 0.2594 | No |
| 18 | Nr4a2 | 3179 | 0.248 | 0.2397 | No |
| 19 | Prodh | 3313 | 0.238 | 0.2419 | No |
| 20 | Epha5 | 3609 | 0.215 | 0.2297 | No |
| 21 | Pdk2 | 3634 | 0.214 | 0.2395 | No |
| 22 | Zbtb16 | 3658 | 0.212 | 0.2493 | No |
| 23 | Lfng | 3691 | 0.210 | 0.2582 | No |
| 24 | Sphk2 | 3879 | 0.192 | 0.2535 | No |
| 25 | Rsad2 | 4716 | 0.126 | 0.1925 | No |
| 26 | Snn | 4941 | 0.113 | 0.1804 | No |
| 27 | Rgs11 | 4969 | 0.110 | 0.1843 | No |
| 28 | Celsr2 | 4996 | 0.110 | 0.1882 | No |
| 29 | Tgm1 | 5037 | 0.109 | 0.1910 | No |
| 30 | Sidt1 | 5114 | 0.104 | 0.1905 | No |
| 31 | Asb7 | 5125 | 0.103 | 0.1953 | No |
| 32 | Slc29a3 | 5396 | 0.081 | 0.1778 | No |
| 33 | Dtnb | 5701 | 0.061 | 0.1564 | No |
| 34 | Ryr2 | 5711 | 0.060 | 0.1589 | No |
| 35 | Mthfr | 5725 | 0.059 | 0.1611 | No |
| 36 | Klk8 | 5917 | 0.046 | 0.1481 | No |
| 37 | Tcf7l1 | 6033 | 0.037 | 0.1408 | No |
| 38 | Tg | 6202 | 0.024 | 0.1284 | No |
| 39 | Fgfr3 | 6356 | 0.013 | 0.1167 | No |
| 40 | Chst2 | 6685 | -0.008 | 0.0905 | No |
| 41 | Cpeb3 | 7184 | -0.044 | 0.0523 | No |
| 42 | Cyp39a1 | 7223 | -0.047 | 0.0518 | No |
| 43 | Selenop | 7447 | -0.066 | 0.0373 | No |
| 44 | Cdkal1 | 7789 | -0.092 | 0.0146 | No |
| 45 | Gprc5c | 7845 | -0.096 | 0.0154 | No |
| 46 | Mx2 | 7901 | -0.100 | 0.0164 | No |
| 47 | Brdt | 7927 | -0.101 | 0.0199 | No |
| 48 | Nr6a1 | 8117 | -0.115 | 0.0109 | No |
| 49 | Vps50 | 8136 | -0.116 | 0.0158 | No |
| 50 | Kmt2d | 8217 | -0.123 | 0.0160 | No |
| 51 | Coq8a | 8470 | -0.144 | 0.0034 | No |
| 52 | Prkn | 8580 | -0.153 | 0.0030 | No |
| 53 | Gtf3c5 | 8736 | -0.162 | -0.0008 | No |
| 54 | P2rx6 | 9333 | -0.215 | -0.0375 | No |
| 55 | Idua | 9845 | -0.259 | -0.0648 | No |
| 56 | Clstn3 | 10050 | -0.278 | -0.0661 | No |
| 57 | Mast3 | 10130 | -0.285 | -0.0569 | No |
| 58 | Mfsd6 | 10232 | -0.296 | -0.0489 | No |
| 59 | Zfp112 | 10277 | -0.299 | -0.0361 | No |
| 60 | Thrb | 10479 | -0.322 | -0.0348 | No |
| 61 | Ptprj | 10700 | -0.348 | -0.0336 | No |
| 62 | Msh5 | 11085 | -0.399 | -0.0429 | No |
| 63 | Bard1 | 11261 | -0.422 | -0.0340 | No |
| 64 | Tnni3 | 11657 | -0.488 | -0.0394 | No |
| 65 | Itgb1bp2 | 11706 | -0.502 | -0.0158 | No |
| 66 | Entpd7 | 12001 | -0.583 | -0.0077 | No |
| 67 | Plag1 | 12174 | -0.676 | 0.0154 | No |