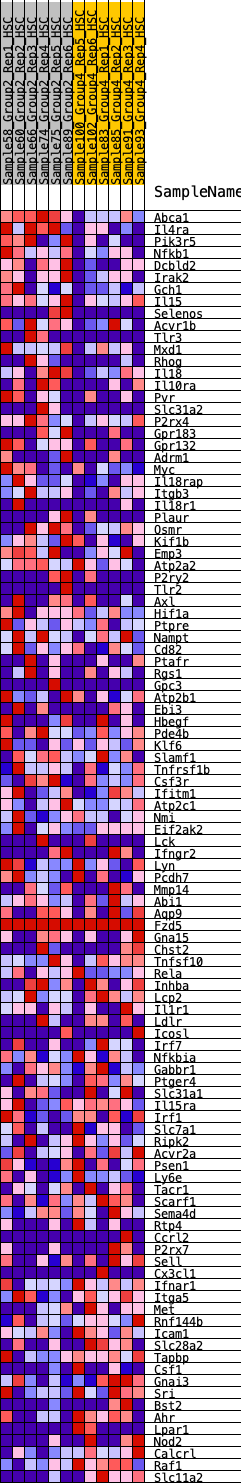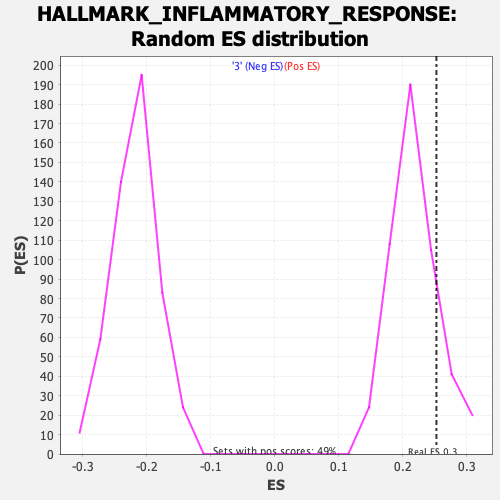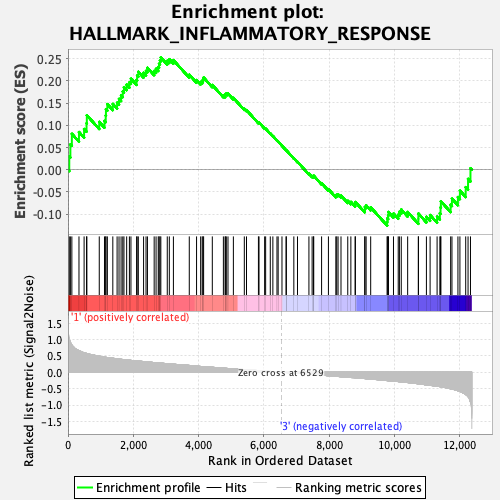
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.25247186 |
| Normalized Enrichment Score (NES) | 1.1637644 |
| Nominal p-value | 0.16803278 |
| FDR q-value | 0.5873458 |
| FWER p-Value | 0.926 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 39 | 1.009 | 0.0300 | Yes |
| 2 | Il4ra | 73 | 0.906 | 0.0571 | Yes |
| 3 | Pik3r5 | 118 | 0.830 | 0.0808 | Yes |
| 4 | Nfkb1 | 336 | 0.658 | 0.0847 | Yes |
| 5 | Dcbld2 | 493 | 0.595 | 0.0916 | Yes |
| 6 | Irak2 | 568 | 0.573 | 0.1044 | Yes |
| 7 | Gch1 | 577 | 0.569 | 0.1224 | Yes |
| 8 | Il15 | 957 | 0.488 | 0.1075 | Yes |
| 9 | Selenos | 1117 | 0.462 | 0.1098 | Yes |
| 10 | Acvr1b | 1155 | 0.456 | 0.1217 | Yes |
| 11 | Tlr3 | 1162 | 0.454 | 0.1362 | Yes |
| 12 | Mxd1 | 1205 | 0.448 | 0.1475 | Yes |
| 13 | Rhog | 1372 | 0.426 | 0.1479 | Yes |
| 14 | Il18 | 1503 | 0.410 | 0.1508 | Yes |
| 15 | Il10ra | 1562 | 0.402 | 0.1593 | Yes |
| 16 | Pvr | 1625 | 0.394 | 0.1672 | Yes |
| 17 | Slc31a2 | 1677 | 0.388 | 0.1758 | Yes |
| 18 | P2rx4 | 1713 | 0.383 | 0.1855 | Yes |
| 19 | Gpr183 | 1796 | 0.376 | 0.1912 | Yes |
| 20 | Gpr132 | 1880 | 0.366 | 0.1965 | Yes |
| 21 | Adrm1 | 1927 | 0.362 | 0.2046 | Yes |
| 22 | Myc | 2100 | 0.344 | 0.2019 | Yes |
| 23 | Il18rap | 2117 | 0.343 | 0.2119 | Yes |
| 24 | Itgb3 | 2153 | 0.340 | 0.2202 | Yes |
| 25 | Il18r1 | 2314 | 0.326 | 0.2179 | Yes |
| 26 | Plaur | 2389 | 0.319 | 0.2223 | Yes |
| 27 | Osmr | 2432 | 0.313 | 0.2292 | Yes |
| 28 | Kif1b | 2638 | 0.296 | 0.2222 | Yes |
| 29 | Emp3 | 2696 | 0.292 | 0.2271 | Yes |
| 30 | Atp2a2 | 2763 | 0.284 | 0.2311 | Yes |
| 31 | P2ry2 | 2788 | 0.281 | 0.2384 | Yes |
| 32 | Tlr2 | 2815 | 0.280 | 0.2454 | Yes |
| 33 | Axl | 2842 | 0.278 | 0.2525 | Yes |
| 34 | Hif1a | 3039 | 0.260 | 0.2450 | No |
| 35 | Ptpre | 3102 | 0.255 | 0.2484 | No |
| 36 | Nampt | 3227 | 0.246 | 0.2464 | No |
| 37 | Cd82 | 3713 | 0.207 | 0.2136 | No |
| 38 | Ptafr | 3936 | 0.186 | 0.2016 | No |
| 39 | Rgs1 | 4061 | 0.176 | 0.1973 | No |
| 40 | Gpc3 | 4108 | 0.174 | 0.1993 | No |
| 41 | Atp2b1 | 4132 | 0.173 | 0.2031 | No |
| 42 | Ebi3 | 4152 | 0.171 | 0.2072 | No |
| 43 | Hbegf | 4418 | 0.151 | 0.1905 | No |
| 44 | Pde4b | 4757 | 0.124 | 0.1670 | No |
| 45 | Klf6 | 4811 | 0.120 | 0.1666 | No |
| 46 | Slamf1 | 4818 | 0.120 | 0.1701 | No |
| 47 | Tnfrsf1b | 4847 | 0.118 | 0.1717 | No |
| 48 | Csf3r | 4898 | 0.115 | 0.1714 | No |
| 49 | Ifitm1 | 5063 | 0.107 | 0.1615 | No |
| 50 | Atp2c1 | 5398 | 0.081 | 0.1370 | No |
| 51 | Nmi | 5465 | 0.077 | 0.1341 | No |
| 52 | Eif2ak2 | 5830 | 0.051 | 0.1061 | No |
| 53 | Lck | 5848 | 0.050 | 0.1063 | No |
| 54 | Ifngr2 | 6020 | 0.038 | 0.0936 | No |
| 55 | Lyn | 6051 | 0.036 | 0.0924 | No |
| 56 | Pcdh7 | 6193 | 0.025 | 0.0817 | No |
| 57 | Mmp14 | 6272 | 0.018 | 0.0759 | No |
| 58 | Abi1 | 6398 | 0.009 | 0.0660 | No |
| 59 | Aqp9 | 6434 | 0.007 | 0.0634 | No |
| 60 | Fzd5 | 6551 | 0.000 | 0.0539 | No |
| 61 | Gna15 | 6676 | -0.007 | 0.0440 | No |
| 62 | Chst2 | 6685 | -0.008 | 0.0436 | No |
| 63 | Tnfsf10 | 6914 | -0.025 | 0.0259 | No |
| 64 | Rela | 7026 | -0.031 | 0.0178 | No |
| 65 | Inhba | 7376 | -0.061 | -0.0087 | No |
| 66 | Lcp2 | 7474 | -0.067 | -0.0144 | No |
| 67 | Il1r1 | 7519 | -0.071 | -0.0156 | No |
| 68 | Ldlr | 7521 | -0.071 | -0.0134 | No |
| 69 | Icosl | 7764 | -0.090 | -0.0301 | No |
| 70 | Irf7 | 7973 | -0.105 | -0.0437 | No |
| 71 | Nfkbia | 8200 | -0.122 | -0.0581 | No |
| 72 | Gabbr1 | 8227 | -0.124 | -0.0562 | No |
| 73 | Ptger4 | 8268 | -0.128 | -0.0552 | No |
| 74 | Slc31a1 | 8358 | -0.135 | -0.0580 | No |
| 75 | Il15ra | 8565 | -0.152 | -0.0699 | No |
| 76 | Irf1 | 8660 | -0.159 | -0.0723 | No |
| 77 | Slc7a1 | 8790 | -0.166 | -0.0774 | No |
| 78 | Ripk2 | 8799 | -0.167 | -0.0725 | No |
| 79 | Acvr2a | 9085 | -0.192 | -0.0895 | No |
| 80 | Psen1 | 9090 | -0.192 | -0.0835 | No |
| 81 | Ly6e | 9127 | -0.195 | -0.0800 | No |
| 82 | Tacr1 | 9267 | -0.210 | -0.0844 | No |
| 83 | Scarf1 | 9772 | -0.253 | -0.1172 | No |
| 84 | Sema4d | 9777 | -0.253 | -0.1092 | No |
| 85 | Rtp4 | 9802 | -0.256 | -0.1027 | No |
| 86 | Ccrl2 | 9811 | -0.258 | -0.0949 | No |
| 87 | P2rx7 | 9965 | -0.271 | -0.0985 | No |
| 88 | Sell | 10110 | -0.284 | -0.1009 | No |
| 89 | Cx3cl1 | 10146 | -0.287 | -0.0944 | No |
| 90 | Ifnar1 | 10203 | -0.292 | -0.0893 | No |
| 91 | Itga5 | 10399 | -0.313 | -0.0949 | No |
| 92 | Met | 10727 | -0.352 | -0.1101 | No |
| 93 | Rnf144b | 10728 | -0.352 | -0.0985 | No |
| 94 | Icam1 | 10973 | -0.384 | -0.1058 | No |
| 95 | Slc28a2 | 11086 | -0.399 | -0.1018 | No |
| 96 | Tapbp | 11300 | -0.426 | -0.1052 | No |
| 97 | Csf1 | 11385 | -0.438 | -0.0976 | No |
| 98 | Gnai3 | 11406 | -0.442 | -0.0848 | No |
| 99 | Sri | 11416 | -0.443 | -0.0709 | No |
| 100 | Bst2 | 11715 | -0.504 | -0.0786 | No |
| 101 | Ahr | 11755 | -0.513 | -0.0650 | No |
| 102 | Lpar1 | 11938 | -0.562 | -0.0613 | No |
| 103 | Nod2 | 11999 | -0.582 | -0.0471 | No |
| 104 | Calcrl | 12176 | -0.677 | -0.0392 | No |
| 105 | Raf1 | 12248 | -0.743 | -0.0206 | No |
| 106 | Slc11a2 | 12322 | -0.909 | 0.0033 | No |