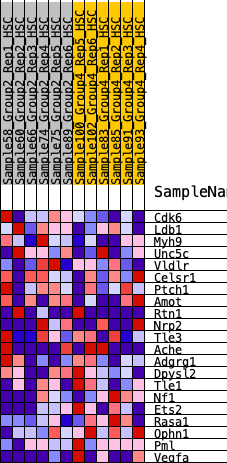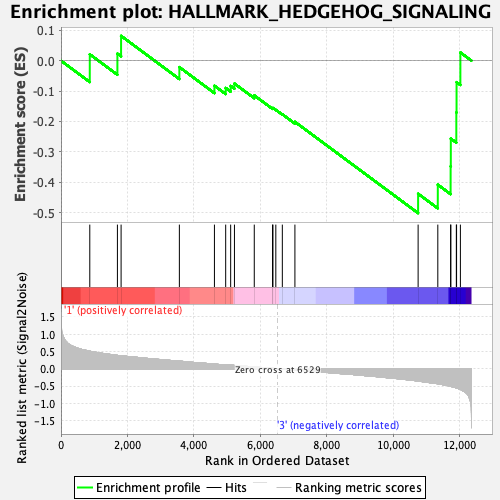
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | -0.5017052 |
| Normalized Enrichment Score (NES) | -1.5353091 |
| Nominal p-value | 0.040462427 |
| FDR q-value | 0.12926665 |
| FWER p-Value | 0.184 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdk6 | 867 | 0.507 | 0.0211 | No |
| 2 | Ldb1 | 1699 | 0.386 | 0.0233 | No |
| 3 | Myh9 | 1810 | 0.374 | 0.0818 | No |
| 4 | Unc5c | 3565 | 0.217 | -0.0212 | No |
| 5 | Vldlr | 4621 | 0.135 | -0.0824 | No |
| 6 | Celsr1 | 4960 | 0.111 | -0.0897 | No |
| 7 | Ptch1 | 5110 | 0.104 | -0.0831 | No |
| 8 | Amot | 5224 | 0.095 | -0.0752 | No |
| 9 | Rtn1 | 5821 | 0.052 | -0.1142 | No |
| 10 | Nrp2 | 6370 | 0.012 | -0.1564 | No |
| 11 | Tle3 | 6384 | 0.011 | -0.1555 | No |
| 12 | Ache | 6470 | 0.004 | -0.1617 | No |
| 13 | Adgrg1 | 6666 | -0.006 | -0.1764 | No |
| 14 | Dpysl2 | 7045 | -0.032 | -0.2011 | No |
| 15 | Tle1 | 10756 | -0.354 | -0.4379 | Yes |
| 16 | Nf1 | 11349 | -0.432 | -0.4080 | Yes |
| 17 | Ets2 | 11735 | -0.508 | -0.3477 | Yes |
| 18 | Rasa1 | 11741 | -0.508 | -0.2564 | Yes |
| 19 | Ophn1 | 11907 | -0.553 | -0.1702 | Yes |
| 20 | Pml | 11914 | -0.555 | -0.0707 | Yes |
| 21 | Vegfa | 12029 | -0.594 | 0.0271 | Yes |