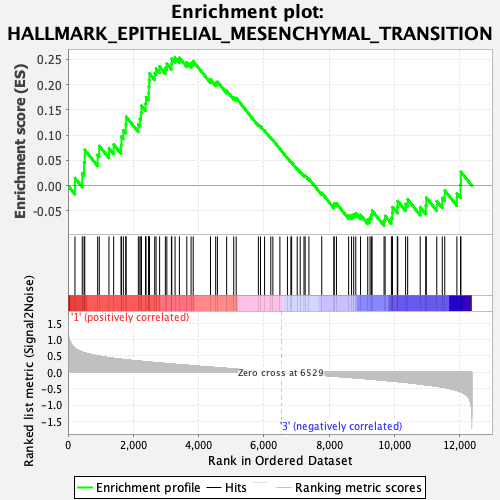
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.25364238 |
| Normalized Enrichment Score (NES) | 1.1134888 |
| Nominal p-value | 0.2237354 |
| FDR q-value | 0.7168491 |
| FWER p-Value | 0.962 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Itgav | 213 | 0.728 | 0.0150 | Yes |
| 2 | Anpep | 436 | 0.615 | 0.0243 | Yes |
| 3 | Dst | 491 | 0.595 | 0.0463 | Yes |
| 4 | Tgfb1 | 514 | 0.589 | 0.0707 | Yes |
| 5 | Lrp1 | 908 | 0.498 | 0.0608 | Yes |
| 6 | Il15 | 957 | 0.488 | 0.0786 | Yes |
| 7 | Cadm1 | 1254 | 0.443 | 0.0742 | Yes |
| 8 | Gadd45b | 1398 | 0.423 | 0.0814 | Yes |
| 9 | Pvr | 1625 | 0.394 | 0.0805 | Yes |
| 10 | Serpinh1 | 1635 | 0.393 | 0.0972 | Yes |
| 11 | Tpm1 | 1695 | 0.386 | 0.1096 | Yes |
| 12 | Vegfc | 1768 | 0.378 | 0.1205 | Yes |
| 13 | Tgfbr3 | 1779 | 0.377 | 0.1364 | Yes |
| 14 | Itgb3 | 2153 | 0.340 | 0.1211 | Yes |
| 15 | Abi3bp | 2200 | 0.335 | 0.1323 | Yes |
| 16 | Lama3 | 2235 | 0.332 | 0.1443 | Yes |
| 17 | Fzd8 | 2248 | 0.332 | 0.1581 | Yes |
| 18 | Ecm1 | 2373 | 0.320 | 0.1622 | Yes |
| 19 | Plaur | 2389 | 0.319 | 0.1751 | Yes |
| 20 | Magee1 | 2468 | 0.310 | 0.1826 | Yes |
| 21 | Lgals1 | 2473 | 0.310 | 0.1960 | Yes |
| 22 | Cap2 | 2486 | 0.310 | 0.2089 | Yes |
| 23 | Fap | 2496 | 0.309 | 0.2219 | Yes |
| 24 | Rhob | 2655 | 0.294 | 0.2221 | Yes |
| 25 | Emp3 | 2696 | 0.292 | 0.2318 | Yes |
| 26 | Timp3 | 2805 | 0.280 | 0.2355 | Yes |
| 27 | Lamc1 | 2980 | 0.266 | 0.2331 | Yes |
| 28 | Jun | 3026 | 0.262 | 0.2411 | Yes |
| 29 | Slit2 | 3166 | 0.249 | 0.2408 | Yes |
| 30 | Calu | 3177 | 0.248 | 0.2511 | Yes |
| 31 | Gpc1 | 3278 | 0.241 | 0.2536 | Yes |
| 32 | Sgcb | 3410 | 0.230 | 0.2532 | No |
| 33 | Plod3 | 3637 | 0.214 | 0.2443 | No |
| 34 | Bgn | 3768 | 0.202 | 0.2427 | No |
| 35 | Col4a1 | 3833 | 0.196 | 0.2462 | No |
| 36 | Qsox1 | 4362 | 0.155 | 0.2101 | No |
| 37 | Sat1 | 4521 | 0.143 | 0.2036 | No |
| 38 | Ecm2 | 4574 | 0.138 | 0.2055 | No |
| 39 | Col4a2 | 4857 | 0.117 | 0.1877 | No |
| 40 | Tpm2 | 5074 | 0.107 | 0.1749 | No |
| 41 | Tpm4 | 5149 | 0.101 | 0.1734 | No |
| 42 | Glipr1 | 5829 | 0.051 | 0.1203 | No |
| 43 | Mmp2 | 5894 | 0.047 | 0.1172 | No |
| 44 | Flna | 6018 | 0.038 | 0.1089 | No |
| 45 | P3h1 | 6209 | 0.024 | 0.0944 | No |
| 46 | Mmp14 | 6272 | 0.018 | 0.0902 | No |
| 47 | Vim | 6487 | 0.003 | 0.0729 | No |
| 48 | Mcm7 | 6718 | -0.010 | 0.0546 | No |
| 49 | Bmp1 | 6824 | -0.018 | 0.0469 | No |
| 50 | Gem | 6847 | -0.019 | 0.0459 | No |
| 51 | Pdgfrb | 7018 | -0.031 | 0.0334 | No |
| 52 | Itga2 | 7109 | -0.038 | 0.0278 | No |
| 53 | Sntb1 | 7225 | -0.047 | 0.0205 | No |
| 54 | Copa | 7261 | -0.050 | 0.0199 | No |
| 55 | Inhba | 7376 | -0.061 | 0.0133 | No |
| 56 | Sfrp1 | 7768 | -0.090 | -0.0146 | No |
| 57 | Colgalt1 | 8140 | -0.117 | -0.0396 | No |
| 58 | Pmepa1 | 8150 | -0.117 | -0.0351 | No |
| 59 | Itgb1 | 8219 | -0.123 | -0.0352 | No |
| 60 | Ppib | 8592 | -0.154 | -0.0586 | No |
| 61 | Fas | 8681 | -0.161 | -0.0587 | No |
| 62 | Eno2 | 8747 | -0.163 | -0.0567 | No |
| 63 | Fstl1 | 8811 | -0.168 | -0.0544 | No |
| 64 | Wipf1 | 8956 | -0.182 | -0.0580 | No |
| 65 | Basp1 | 9177 | -0.201 | -0.0670 | No |
| 66 | Col1a2 | 9247 | -0.208 | -0.0634 | No |
| 67 | Capg | 9288 | -0.212 | -0.0573 | No |
| 68 | Plod1 | 9310 | -0.213 | -0.0495 | No |
| 69 | Plod2 | 9676 | -0.244 | -0.0683 | No |
| 70 | Tgfbi | 9709 | -0.248 | -0.0599 | No |
| 71 | Col16a1 | 9901 | -0.263 | -0.0638 | No |
| 72 | Tnfaip3 | 9927 | -0.267 | -0.0540 | No |
| 73 | Gja1 | 9933 | -0.268 | -0.0425 | No |
| 74 | Vcan | 10082 | -0.282 | -0.0420 | No |
| 75 | Notch2 | 10096 | -0.282 | -0.0305 | No |
| 76 | Igfbp4 | 10336 | -0.307 | -0.0363 | No |
| 77 | Itga5 | 10399 | -0.313 | -0.0274 | No |
| 78 | Fuca1 | 10783 | -0.358 | -0.0427 | No |
| 79 | Mylk | 10953 | -0.382 | -0.0395 | No |
| 80 | Fermt2 | 10969 | -0.383 | -0.0237 | No |
| 81 | Sdc4 | 11290 | -0.426 | -0.0308 | No |
| 82 | Cd44 | 11459 | -0.452 | -0.0244 | No |
| 83 | Thbs1 | 11535 | -0.464 | -0.0099 | No |
| 84 | Fbn1 | 11906 | -0.552 | -0.0155 | No |
| 85 | Tgm2 | 12017 | -0.588 | 0.0017 | No |
| 86 | Vegfa | 12029 | -0.594 | 0.0272 | No |

