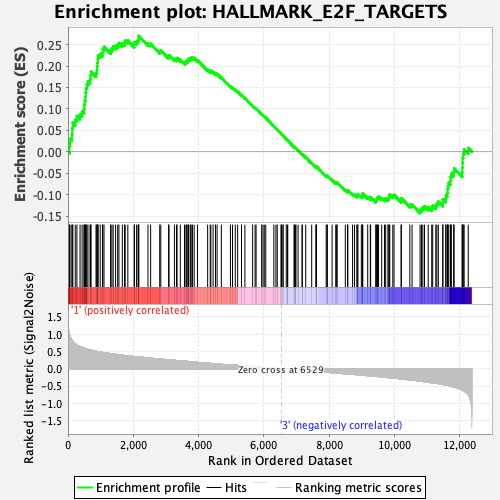
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_E2F_TARGETS |
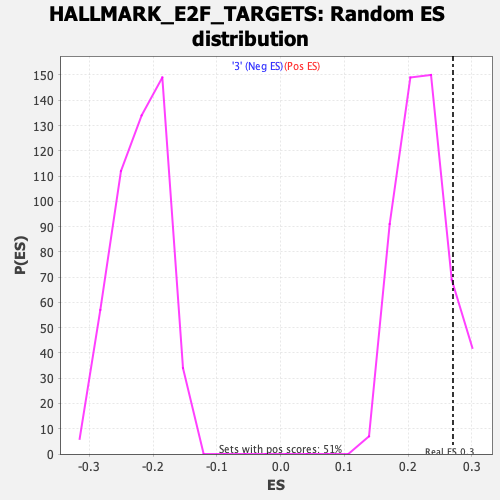
| Enrichment Score (ES) | 0.27030978 |
| Normalized Enrichment Score (NES) | 1.2091902 |
| Nominal p-value | 0.13976377 |
| FDR q-value | 0.5478132 |
| FWER p-Value | 0.867 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Xrcc6 | 45 | 0.988 | 0.0144 | Yes |
| 2 | Smc1a | 62 | 0.936 | 0.0302 | Yes |
| 3 | Mlh1 | 120 | 0.826 | 0.0406 | Yes |
| 4 | Bub1b | 129 | 0.815 | 0.0548 | Yes |
| 5 | Hus1 | 148 | 0.791 | 0.0678 | Yes |
| 6 | Rfc1 | 222 | 0.723 | 0.0750 | Yes |
| 7 | Msh2 | 272 | 0.688 | 0.0836 | Yes |
| 8 | Ak2 | 369 | 0.644 | 0.0875 | Yes |
| 9 | Paics | 431 | 0.616 | 0.0937 | Yes |
| 10 | Rad1 | 489 | 0.596 | 0.0999 | Yes |
| 11 | Birc5 | 497 | 0.594 | 0.1102 | Yes |
| 12 | Rfc3 | 515 | 0.589 | 0.1196 | Yes |
| 13 | Tacc3 | 535 | 0.584 | 0.1287 | Yes |
| 14 | Ipo7 | 544 | 0.580 | 0.1387 | Yes |
| 15 | Chek2 | 553 | 0.578 | 0.1486 | Yes |
| 16 | Hnrnpd | 576 | 0.570 | 0.1572 | Yes |
| 17 | Nop56 | 611 | 0.561 | 0.1647 | Yes |
| 18 | Rnaseh2a | 675 | 0.546 | 0.1695 | Yes |
| 19 | Eed | 691 | 0.542 | 0.1781 | Yes |
| 20 | Pcna | 706 | 0.540 | 0.1869 | Yes |
| 21 | Lig1 | 858 | 0.509 | 0.1838 | Yes |
| 22 | Psmc3ip | 885 | 0.503 | 0.1908 | Yes |
| 23 | Exosc8 | 889 | 0.502 | 0.1997 | Yes |
| 24 | Srsf1 | 902 | 0.499 | 0.2079 | Yes |
| 25 | Tra2b | 914 | 0.497 | 0.2161 | Yes |
| 26 | Mcm6 | 919 | 0.495 | 0.2248 | Yes |
| 27 | Hmgb2 | 986 | 0.485 | 0.2282 | Yes |
| 28 | Stmn1 | 1056 | 0.473 | 0.2312 | Yes |
| 29 | Tfrc | 1063 | 0.472 | 0.2393 | Yes |
| 30 | Pds5b | 1102 | 0.465 | 0.2447 | Yes |
| 31 | Pa2g4 | 1303 | 0.436 | 0.2363 | Yes |
| 32 | Spag5 | 1343 | 0.431 | 0.2409 | Yes |
| 33 | Ctcf | 1383 | 0.424 | 0.2455 | Yes |
| 34 | Cdk1 | 1457 | 0.415 | 0.2471 | Yes |
| 35 | Rpa2 | 1521 | 0.408 | 0.2494 | Yes |
| 36 | Racgap1 | 1559 | 0.403 | 0.2537 | Yes |
| 37 | Cdc20 | 1663 | 0.390 | 0.2524 | Yes |
| 38 | Kif4 | 1732 | 0.381 | 0.2538 | Yes |
| 39 | Brms1l | 1750 | 0.380 | 0.2593 | Yes |
| 40 | Usp1 | 1832 | 0.372 | 0.2595 | Yes |
| 41 | H2az1 | 2025 | 0.352 | 0.2502 | Yes |
| 42 | Rbbp7 | 2034 | 0.352 | 0.2559 | Yes |
| 43 | Myc | 2100 | 0.344 | 0.2569 | Yes |
| 44 | Dnmt1 | 2145 | 0.341 | 0.2595 | Yes |
| 45 | Cbx5 | 2163 | 0.339 | 0.2643 | Yes |
| 46 | Cdkn1b | 2166 | 0.338 | 0.2703 | Yes |
| 47 | Nup107 | 2449 | 0.312 | 0.2529 | No |
| 48 | Syncrip | 2530 | 0.306 | 0.2519 | No |
| 49 | Mthfd2 | 2808 | 0.280 | 0.2343 | No |
| 50 | Smc3 | 2841 | 0.279 | 0.2367 | No |
| 51 | Top2a | 3079 | 0.256 | 0.2220 | No |
| 52 | Hmmr | 3093 | 0.255 | 0.2256 | No |
| 53 | Pms2 | 3258 | 0.243 | 0.2166 | No |
| 54 | Mcm3 | 3321 | 0.238 | 0.2158 | No |
| 55 | Nasp | 3339 | 0.236 | 0.2187 | No |
| 56 | Zw10 | 3438 | 0.228 | 0.2149 | No |
| 57 | Pttg1 | 3567 | 0.217 | 0.2083 | No |
| 58 | Ube2s | 3614 | 0.215 | 0.2085 | No |
| 59 | Cdc25a | 3623 | 0.215 | 0.2117 | No |
| 60 | Lmnb1 | 3667 | 0.211 | 0.2121 | No |
| 61 | Gins3 | 3687 | 0.210 | 0.2143 | No |
| 62 | Gins1 | 3700 | 0.209 | 0.2172 | No |
| 63 | Ilf3 | 3755 | 0.203 | 0.2164 | No |
| 64 | Ing3 | 3769 | 0.202 | 0.2191 | No |
| 65 | Tipin | 3810 | 0.198 | 0.2194 | No |
| 66 | Gins4 | 3862 | 0.194 | 0.2188 | No |
| 67 | Mms22l | 3964 | 0.184 | 0.2138 | No |
| 68 | Cse1l | 4275 | 0.162 | 0.1914 | No |
| 69 | Nme1 | 4354 | 0.156 | 0.1878 | No |
| 70 | Slbp | 4373 | 0.154 | 0.1891 | No |
| 71 | Luc7l3 | 4441 | 0.149 | 0.1864 | No |
| 72 | Srsf2 | 4515 | 0.144 | 0.1830 | No |
| 73 | Mki67 | 4565 | 0.139 | 0.1815 | No |
| 74 | Ubr7 | 4694 | 0.128 | 0.1733 | No |
| 75 | Depdc1a | 4973 | 0.110 | 0.1525 | No |
| 76 | Ddx39a | 5036 | 0.109 | 0.1495 | No |
| 77 | Cenpe | 5121 | 0.103 | 0.1444 | No |
| 78 | Asf1a | 5194 | 0.097 | 0.1403 | No |
| 79 | Rpa3 | 5312 | 0.088 | 0.1323 | No |
| 80 | Nap1l1 | 5416 | 0.080 | 0.1253 | No |
| 81 | Ran | 5653 | 0.064 | 0.1071 | No |
| 82 | Dlgap5 | 5728 | 0.059 | 0.1021 | No |
| 83 | Suv39h1 | 5758 | 0.056 | 0.1008 | No |
| 84 | Prps1 | 5928 | 0.044 | 0.0877 | No |
| 85 | Ccp110 | 5970 | 0.042 | 0.0851 | No |
| 86 | Lsm8 | 6009 | 0.039 | 0.0827 | No |
| 87 | Hmgb3 | 6055 | 0.035 | 0.0796 | No |
| 88 | Shmt1 | 6302 | 0.016 | 0.0597 | No |
| 89 | Dclre1b | 6366 | 0.012 | 0.0548 | No |
| 90 | Snrpb | 6406 | 0.009 | 0.0518 | No |
| 91 | Ncapd2 | 6520 | 0.001 | 0.0425 | No |
| 92 | Ccnb2 | 6537 | 0.000 | 0.0412 | No |
| 93 | Kif18b | 6556 | 0.000 | 0.0397 | No |
| 94 | Tubb5 | 6591 | -0.001 | 0.0369 | No |
| 95 | Pole | 6682 | -0.008 | 0.0297 | No |
| 96 | Mcm7 | 6718 | -0.010 | 0.0270 | No |
| 97 | Dut | 6728 | -0.011 | 0.0265 | No |
| 98 | Prkdc | 6919 | -0.025 | 0.0113 | No |
| 99 | Stag1 | 6936 | -0.026 | 0.0105 | No |
| 100 | Mcm4 | 6969 | -0.029 | 0.0084 | No |
| 101 | Cdkn2c | 6978 | -0.029 | 0.0083 | No |
| 102 | Diaph3 | 7044 | -0.032 | 0.0036 | No |
| 103 | Rad21 | 7167 | -0.042 | -0.0057 | No |
| 104 | Mcm2 | 7173 | -0.043 | -0.0053 | No |
| 105 | Lbr | 7278 | -0.052 | -0.0129 | No |
| 106 | Nup205 | 7461 | -0.067 | -0.0266 | No |
| 107 | Nolc1 | 7593 | -0.077 | -0.0360 | No |
| 108 | Lyar | 7594 | -0.077 | -0.0346 | No |
| 109 | Timeless | 7609 | -0.078 | -0.0343 | No |
| 110 | Mybl2 | 7904 | -0.100 | -0.0566 | No |
| 111 | Jpt1 | 7931 | -0.102 | -0.0569 | No |
| 112 | Nup153 | 7940 | -0.102 | -0.0557 | No |
| 113 | Anp32e | 8086 | -0.113 | -0.0655 | No |
| 114 | Wee1 | 8197 | -0.121 | -0.0723 | No |
| 115 | Trp53 | 8225 | -0.124 | -0.0723 | No |
| 116 | Prdx4 | 8240 | -0.125 | -0.0711 | No |
| 117 | Orc6 | 8490 | -0.146 | -0.0889 | No |
| 118 | Hells | 8558 | -0.152 | -0.0916 | No |
| 119 | Mre11a | 8568 | -0.152 | -0.0896 | No |
| 120 | Cks2 | 8712 | -0.161 | -0.0984 | No |
| 121 | Ssrp1 | 8771 | -0.164 | -0.1001 | No |
| 122 | Atad2 | 8847 | -0.172 | -0.1031 | No |
| 123 | Cit | 8852 | -0.172 | -0.1003 | No |
| 124 | Tbrg4 | 8877 | -0.174 | -0.0991 | No |
| 125 | Dck | 8981 | -0.184 | -0.1042 | No |
| 126 | Pola2 | 9012 | -0.187 | -0.1032 | No |
| 127 | Spc25 | 9019 | -0.188 | -0.1003 | No |
| 128 | Rrm2 | 9024 | -0.188 | -0.0972 | No |
| 129 | Prim2 | 9176 | -0.201 | -0.1059 | No |
| 130 | E2f8 | 9255 | -0.209 | -0.1085 | No |
| 131 | Nbn | 9265 | -0.210 | -0.1054 | No |
| 132 | Rpa1 | 9421 | -0.221 | -0.1141 | No |
| 133 | Cenpm | 9441 | -0.222 | -0.1116 | No |
| 134 | Nudt21 | 9461 | -0.223 | -0.1090 | No |
| 135 | Pan2 | 9476 | -0.225 | -0.1061 | No |
| 136 | Psip1 | 9508 | -0.227 | -0.1045 | No |
| 137 | Ranbp1 | 9604 | -0.237 | -0.1079 | No |
| 138 | Pold3 | 9689 | -0.246 | -0.1103 | No |
| 139 | Ccne1 | 9726 | -0.250 | -0.1087 | No |
| 140 | Dscc1 | 9795 | -0.256 | -0.1096 | No |
| 141 | Cdc25b | 9826 | -0.259 | -0.1074 | No |
| 142 | Tk1 | 9829 | -0.259 | -0.1028 | No |
| 143 | Xpo1 | 9851 | -0.260 | -0.0998 | No |
| 144 | Plk4 | 9944 | -0.268 | -0.1024 | No |
| 145 | Rad50 | 9982 | -0.272 | -0.1005 | No |
| 146 | Mad2l1 | 10198 | -0.291 | -0.1128 | No |
| 147 | Ezh2 | 10205 | -0.292 | -0.1080 | No |
| 148 | Mcm5 | 10466 | -0.321 | -0.1235 | No |
| 149 | Rad51ap1 | 10534 | -0.328 | -0.1230 | No |
| 150 | Donson | 10779 | -0.358 | -0.1364 | No |
| 151 | Chek1 | 10822 | -0.363 | -0.1333 | No |
| 152 | Brca1 | 10863 | -0.369 | -0.1298 | No |
| 153 | Tmpo | 10914 | -0.377 | -0.1270 | No |
| 154 | Rfc2 | 11028 | -0.390 | -0.1291 | No |
| 155 | Tubg1 | 11136 | -0.405 | -0.1305 | No |
| 156 | Pole4 | 11166 | -0.408 | -0.1254 | No |
| 157 | Bard1 | 11261 | -0.422 | -0.1255 | No |
| 158 | Aurka | 11293 | -0.426 | -0.1202 | No |
| 159 | Cdca8 | 11337 | -0.429 | -0.1159 | No |
| 160 | Smc4 | 11475 | -0.455 | -0.1188 | No |
| 161 | Wdr90 | 11476 | -0.455 | -0.1105 | No |
| 162 | Smc6 | 11565 | -0.469 | -0.1092 | No |
| 163 | Ppm1d | 11574 | -0.471 | -0.1012 | No |
| 164 | Phf5a | 11614 | -0.480 | -0.0957 | No |
| 165 | Ube2t | 11621 | -0.481 | -0.0874 | No |
| 166 | Dctpp1 | 11633 | -0.484 | -0.0794 | No |
| 167 | Pnn | 11658 | -0.488 | -0.0725 | No |
| 168 | Gspt1 | 11710 | -0.503 | -0.0675 | No |
| 169 | Eif2s1 | 11713 | -0.504 | -0.0584 | No |
| 170 | Cnot9 | 11737 | -0.508 | -0.0510 | No |
| 171 | Pold2 | 11807 | -0.524 | -0.0471 | No |
| 172 | Dek | 11821 | -0.528 | -0.0385 | No |
| 173 | Ppp1r8 | 12067 | -0.615 | -0.0474 | No |
| 174 | Trip13 | 12074 | -0.618 | -0.0366 | No |
| 175 | Cdk4 | 12079 | -0.621 | -0.0256 | No |
| 176 | H2ax | 12084 | -0.624 | -0.0145 | No |
| 177 | Pold1 | 12101 | -0.633 | -0.0042 | No |
| 178 | Orc2 | 12128 | -0.648 | 0.0055 | No |
| 179 | Brca2 | 12254 | -0.750 | 0.0089 | No |