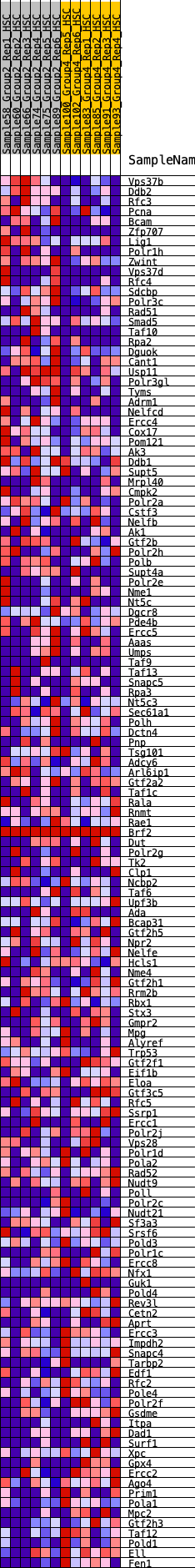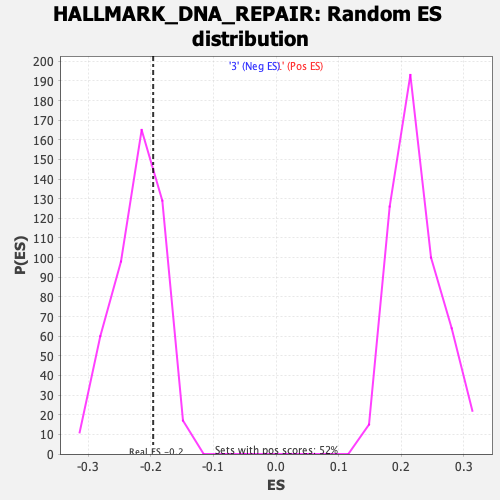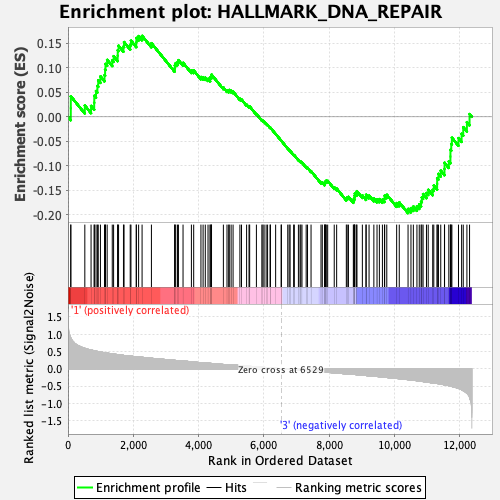
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | -0.19628347 |
| Normalized Enrichment Score (NES) | -0.8878094 |
| Nominal p-value | 0.71666664 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vps37b | 85 | 0.889 | 0.0175 | No |
| 2 | Ddb2 | 86 | 0.886 | 0.0418 | No |
| 3 | Rfc3 | 515 | 0.589 | 0.0230 | No |
| 4 | Pcna | 706 | 0.540 | 0.0223 | No |
| 5 | Bcam | 802 | 0.522 | 0.0288 | No |
| 6 | Zfp707 | 805 | 0.521 | 0.0430 | No |
| 7 | Lig1 | 858 | 0.509 | 0.0527 | No |
| 8 | Polr1h | 900 | 0.499 | 0.0631 | No |
| 9 | Zwint | 926 | 0.493 | 0.0746 | No |
| 10 | Vps37d | 993 | 0.483 | 0.0824 | No |
| 11 | Rfc4 | 1118 | 0.462 | 0.0850 | No |
| 12 | Sdcbp | 1136 | 0.459 | 0.0962 | No |
| 13 | Polr3c | 1148 | 0.457 | 0.1078 | No |
| 14 | Rad51 | 1199 | 0.449 | 0.1161 | No |
| 15 | Smad5 | 1355 | 0.429 | 0.1152 | No |
| 16 | Taf10 | 1396 | 0.423 | 0.1235 | No |
| 17 | Rpa2 | 1521 | 0.408 | 0.1246 | No |
| 18 | Dguok | 1523 | 0.408 | 0.1357 | No |
| 19 | Cant1 | 1548 | 0.405 | 0.1449 | No |
| 20 | Usp11 | 1698 | 0.386 | 0.1433 | No |
| 21 | Polr3gl | 1716 | 0.383 | 0.1524 | No |
| 22 | Tyms | 1905 | 0.363 | 0.1470 | No |
| 23 | Adrm1 | 1927 | 0.362 | 0.1552 | No |
| 24 | Nelfcd | 2090 | 0.345 | 0.1515 | No |
| 25 | Ercc4 | 2093 | 0.345 | 0.1608 | No |
| 26 | Cox17 | 2162 | 0.339 | 0.1645 | No |
| 27 | Pom121 | 2268 | 0.330 | 0.1650 | No |
| 28 | Ak3 | 2554 | 0.304 | 0.1500 | No |
| 29 | Ddb1 | 3265 | 0.242 | 0.0986 | No |
| 30 | Supt5 | 3271 | 0.242 | 0.1049 | No |
| 31 | Mrpl40 | 3297 | 0.239 | 0.1094 | No |
| 32 | Cmpk2 | 3351 | 0.235 | 0.1115 | No |
| 33 | Polr2a | 3381 | 0.232 | 0.1155 | No |
| 34 | Cstf3 | 3522 | 0.221 | 0.1101 | No |
| 35 | Nelfb | 3778 | 0.201 | 0.0948 | No |
| 36 | Ak1 | 3848 | 0.195 | 0.0945 | No |
| 37 | Gtf2b | 4070 | 0.175 | 0.0812 | No |
| 38 | Polr2h | 4136 | 0.173 | 0.0807 | No |
| 39 | Polb | 4203 | 0.168 | 0.0799 | No |
| 40 | Supt4a | 4287 | 0.161 | 0.0775 | No |
| 41 | Polr2e | 4352 | 0.156 | 0.0766 | No |
| 42 | Nme1 | 4354 | 0.156 | 0.0807 | No |
| 43 | Nt5c | 4390 | 0.152 | 0.0821 | No |
| 44 | Dgcr8 | 4394 | 0.152 | 0.0860 | No |
| 45 | Pde4b | 4757 | 0.124 | 0.0598 | No |
| 46 | Ercc5 | 4868 | 0.116 | 0.0540 | No |
| 47 | Aaas | 4921 | 0.114 | 0.0528 | No |
| 48 | Umps | 4936 | 0.113 | 0.0548 | No |
| 49 | Taf9 | 4994 | 0.110 | 0.0532 | No |
| 50 | Taf13 | 5054 | 0.108 | 0.0513 | No |
| 51 | Snapc5 | 5263 | 0.092 | 0.0368 | No |
| 52 | Rpa3 | 5312 | 0.088 | 0.0353 | No |
| 53 | Nt5c3 | 5468 | 0.077 | 0.0247 | No |
| 54 | Sec61a1 | 5542 | 0.071 | 0.0207 | No |
| 55 | Polh | 5562 | 0.070 | 0.0211 | No |
| 56 | Dctn4 | 5768 | 0.055 | 0.0058 | No |
| 57 | Pnp | 5935 | 0.044 | -0.0066 | No |
| 58 | Tsg101 | 5967 | 0.042 | -0.0079 | No |
| 59 | Adcy6 | 6003 | 0.039 | -0.0097 | No |
| 60 | Arl6ip1 | 6066 | 0.034 | -0.0139 | No |
| 61 | Gtf2a2 | 6110 | 0.031 | -0.0165 | No |
| 62 | Taf1c | 6187 | 0.025 | -0.0220 | No |
| 63 | Rala | 6198 | 0.024 | -0.0222 | No |
| 64 | Rnmt | 6357 | 0.013 | -0.0348 | No |
| 65 | Rae1 | 6525 | 0.000 | -0.0484 | No |
| 66 | Brf2 | 6535 | 0.000 | -0.0492 | No |
| 67 | Dut | 6728 | -0.011 | -0.0645 | No |
| 68 | Polr2g | 6770 | -0.014 | -0.0675 | No |
| 69 | Tk2 | 6800 | -0.017 | -0.0694 | No |
| 70 | Clp1 | 6908 | -0.024 | -0.0775 | No |
| 71 | Ncbp2 | 6929 | -0.026 | -0.0784 | No |
| 72 | Taf6 | 7054 | -0.033 | -0.0877 | No |
| 73 | Upf3b | 7103 | -0.037 | -0.0906 | No |
| 74 | Ada | 7128 | -0.039 | -0.0915 | No |
| 75 | Bcap31 | 7163 | -0.042 | -0.0931 | No |
| 76 | Gtf2h5 | 7292 | -0.054 | -0.1021 | No |
| 77 | Npr2 | 7333 | -0.057 | -0.1038 | No |
| 78 | Nelfe | 7441 | -0.065 | -0.1108 | No |
| 79 | Hcls1 | 7746 | -0.089 | -0.1332 | No |
| 80 | Nme4 | 7785 | -0.092 | -0.1338 | No |
| 81 | Gtf2h1 | 7856 | -0.096 | -0.1369 | No |
| 82 | Rrm2b | 7858 | -0.096 | -0.1343 | No |
| 83 | Rbx1 | 7878 | -0.098 | -0.1332 | No |
| 84 | Stx3 | 7887 | -0.099 | -0.1311 | No |
| 85 | Gmpr2 | 7907 | -0.100 | -0.1299 | No |
| 86 | Mpg | 7950 | -0.103 | -0.1305 | No |
| 87 | Alyref | 8151 | -0.117 | -0.1437 | No |
| 88 | Trp53 | 8225 | -0.124 | -0.1463 | No |
| 89 | Gtf2f1 | 8521 | -0.149 | -0.1663 | No |
| 90 | Eif1b | 8546 | -0.151 | -0.1641 | No |
| 91 | Eloa | 8586 | -0.154 | -0.1631 | No |
| 92 | Gtf3c5 | 8736 | -0.162 | -0.1709 | No |
| 93 | Rfc5 | 8751 | -0.163 | -0.1675 | No |
| 94 | Ssrp1 | 8771 | -0.164 | -0.1646 | No |
| 95 | Ercc1 | 8775 | -0.165 | -0.1603 | No |
| 96 | Polr2j | 8783 | -0.165 | -0.1563 | No |
| 97 | Vps28 | 8840 | -0.171 | -0.1562 | No |
| 98 | Polr1d | 8842 | -0.171 | -0.1516 | No |
| 99 | Pola2 | 9012 | -0.187 | -0.1602 | No |
| 100 | Rad52 | 9125 | -0.195 | -0.1640 | No |
| 101 | Nudt9 | 9126 | -0.195 | -0.1587 | No |
| 102 | Poll | 9217 | -0.205 | -0.1604 | No |
| 103 | Polr2c | 9364 | -0.215 | -0.1665 | No |
| 104 | Nudt21 | 9461 | -0.223 | -0.1682 | No |
| 105 | Sf3a3 | 9533 | -0.230 | -0.1677 | No |
| 106 | Srsf6 | 9626 | -0.239 | -0.1686 | No |
| 107 | Pold3 | 9689 | -0.246 | -0.1669 | No |
| 108 | Polr1c | 9696 | -0.247 | -0.1606 | No |
| 109 | Ercc8 | 9760 | -0.253 | -0.1589 | No |
| 110 | Nfx1 | 10062 | -0.280 | -0.1758 | No |
| 111 | Guk1 | 10142 | -0.287 | -0.1744 | No |
| 112 | Pold4 | 10411 | -0.315 | -0.1876 | Yes |
| 113 | Rev3l | 10500 | -0.325 | -0.1859 | Yes |
| 114 | Cetn2 | 10573 | -0.332 | -0.1827 | Yes |
| 115 | Aprt | 10683 | -0.346 | -0.1821 | Yes |
| 116 | Ercc3 | 10754 | -0.354 | -0.1781 | Yes |
| 117 | Impdh2 | 10810 | -0.361 | -0.1727 | Yes |
| 118 | Snapc4 | 10832 | -0.365 | -0.1644 | Yes |
| 119 | Tarbp2 | 10871 | -0.370 | -0.1573 | Yes |
| 120 | Edf1 | 10975 | -0.384 | -0.1552 | Yes |
| 121 | Rfc2 | 11028 | -0.390 | -0.1487 | Yes |
| 122 | Pole4 | 11166 | -0.408 | -0.1487 | Yes |
| 123 | Polr2f | 11199 | -0.412 | -0.1400 | Yes |
| 124 | Gsdme | 11301 | -0.426 | -0.1366 | Yes |
| 125 | Itpa | 11303 | -0.427 | -0.1249 | Yes |
| 126 | Dad1 | 11342 | -0.430 | -0.1162 | Yes |
| 127 | Surf1 | 11412 | -0.443 | -0.1097 | Yes |
| 128 | Xpc | 11526 | -0.463 | -0.1062 | Yes |
| 129 | Gpx4 | 11529 | -0.464 | -0.0937 | Yes |
| 130 | Ercc2 | 11659 | -0.488 | -0.0908 | Yes |
| 131 | Ago4 | 11708 | -0.502 | -0.0809 | Yes |
| 132 | Prim1 | 11709 | -0.502 | -0.0671 | Yes |
| 133 | Pola1 | 11734 | -0.508 | -0.0551 | Yes |
| 134 | Mpc2 | 11753 | -0.512 | -0.0425 | Yes |
| 135 | Gtf2h3 | 11955 | -0.568 | -0.0434 | Yes |
| 136 | Taf12 | 12051 | -0.608 | -0.0344 | Yes |
| 137 | Pold1 | 12101 | -0.633 | -0.0211 | Yes |
| 138 | Ell | 12213 | -0.704 | -0.0108 | Yes |
| 139 | Fen1 | 12294 | -0.837 | 0.0056 | Yes |